It is reported by the UN Convention to Combat Desertification (UNCCD COP15) that about 40% of the land area not covered by ice worldwide has been subjected to soil degradation, becoming one of the most serious threats to food security and soil biodiversity. The soil microbiome plays an integral role in maintaining food supply and sustainable agriculture for a continuously growing global human population. One of their major roles is to create a protective barrier to face the entrance of soil-borne pathogens and infection by nutrient competition or substance antagonism. This inherent disease-suppressive capacity of soils driven by microbes can result in a reduction in the susceptibility to the disease of a next generation of plants, which is also known as “soil-borne legacy”. Yet, till now we did not understand what the major biotic and abiotic modulators of soil disease-suppression are, and experimental work supporting this knowledge is lacking.
To explore the influence of the soil microbiome as modulator of plant diseases and soil disease-suppression, we conducted a large-scale field investigation across southeast China (extending a rough gradient of latitude from 28.10° to 28.90° N and longitude from 115.00° to 116.96° E) using peanut as our model system. Here, red soils have developed on quaternary red clay [Udic Ferrosol, FAO (1998)] with zonal distribution, and the weather conditions of these fields were comparable. This soil is low in soil organic matters and fertility, the crop types that can be grown are limited. These aforementioned conditions provided an ideal practice to investigate between soil degradation in arable fields and plant diseases, as both plant varieties and soil types are important factors closely related to plant resistance and soil property changes, respectively.
The sampling took place in late July 2018, shortly before the sprout of root disease of peanut (Figure 1). This time is around the peak of summer, with the daytime temperature reaching up to 40 °C. After receiving consent from the farmers, we located each of the sample fields, and recorded the cultivation history of the peanut fields. So, we would extend our gratitude to all our team members, some of whom even suffered from sunstroke, and to the guides who facilitated efficient field work.

Figure 1. (a) A view of field investigation and sampling (Photograph: Xiaogang Li), (b) the healthy peanut in a field (Photograph: Xiaogang Li), (c) Severe plant disease (peanut root rot) in acidified soils (Photograph: Xiaogang Li)
After that, we established detailed work schedules and standardized the workflow when proceeding to the lab experiments (Figure 2). We determined a broad range of the sampled soil properties (16 metrics), and soils supporting disease-suppressive capacities using Fusarium sp. ACCC 36194, a model peanut root rot pathogen both in vitro and in vivo. To discern the association of soil properties and microbiomes with plant status, we first run simple statistical models and stepwise regression aiming at quantify the capacity of our soil attributes to explain disease. Collectively, these data suggest that soil health in arable fields of the hilly red soil region is primarily reflected by pH, with acidification markedly affecting host plant status. After that, we focused on the potential role of soil acidification-microbiome interactions in mediating plant disease severity, highlighting the microbial community assemblies responsible for pathogen invasion. Moreover, we employed shotgun metagenomic sequencing and untargeted metabolomics to explore microbial function in defensing plant health by analyzing pathways enriched in different KO functional categories and identifying specific metabolites emitted by soil microbiome from acidified soils of contrasting degrees.
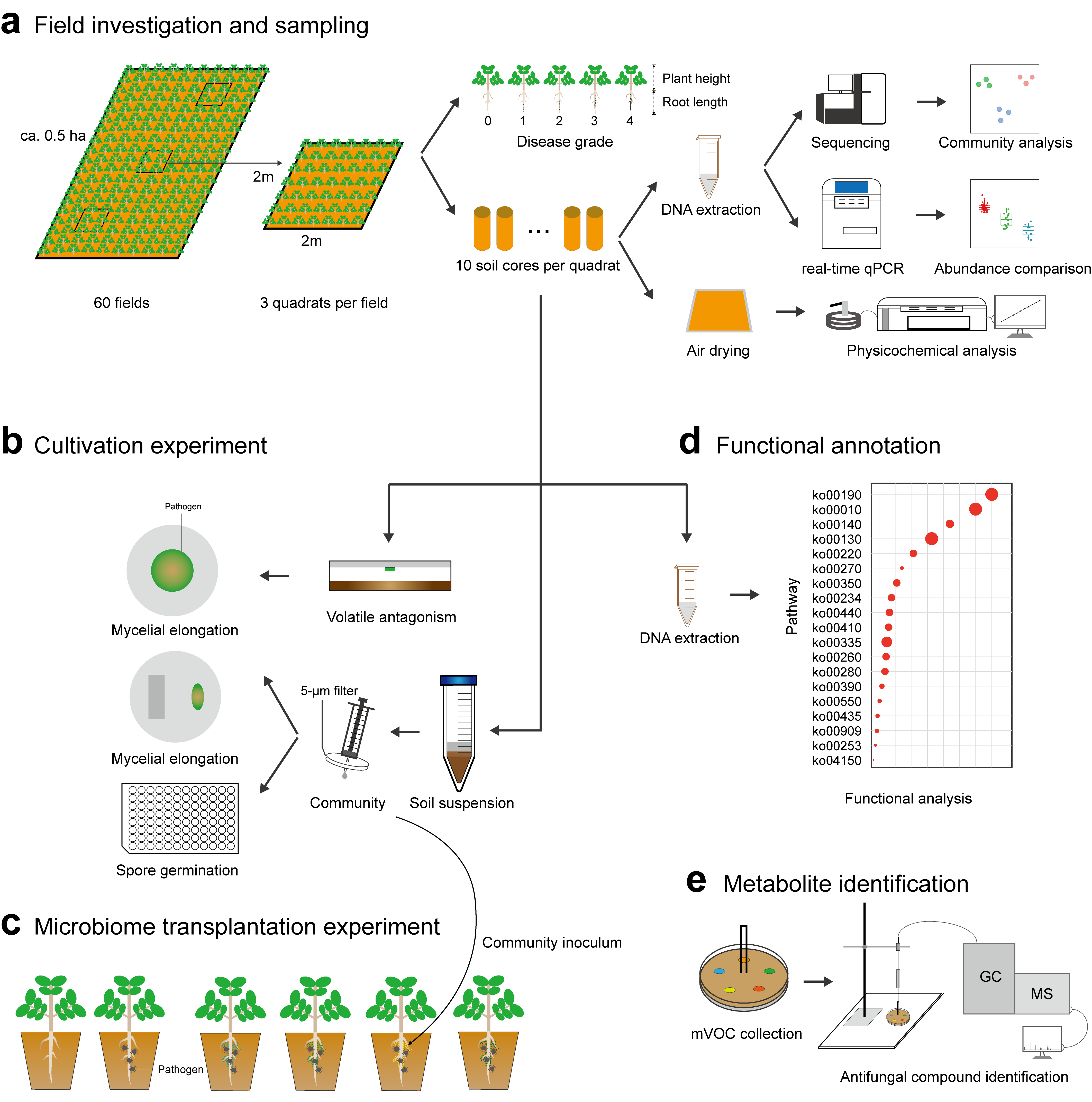
Figure 2. (a) Field investigation and sampling. Three quadrats of 2m × 2m were selected randomly for sampling in each field. Plant disease severity was determined, and soil cores homogenized and transported to the lab for DNA extraction and soil physicochemical analysis. (b) Cultivation experiment. Fungal mycelium of the pathogen Fusarium sp. was exposed to soil microbial volatiles. Pathogen mycelium and spores were co-cultured with bacterial communities on agar and in suspensions to reveal the suppressive effect of bacterial communities on pathogen growth and reproduction. (c) Microbiome transplantation experiment. The bacterial suspensions obtained from soil samples were inoculated in sterilized vermiculite for growth of peanut seedlings. Sterile deionized water was used in controls. (d) Functional annotation. Metagenomic sequencing of 12 soil communities at pH of 4.0-4.5, 4.5-5.0, and 5.0-6.0 was conducted to disclose the functional profiles of the soil microbiome. (e) Metabolite identification. Volatile compounds produced by soil microbial communities were identified by GC-MS.
Our study highlights the significant role of soil acidification in worsening plant health, in comparison to other physiochemical properties of soil. Soil acidification has led to the declines of abundance and diversity of bacterial community, whereas induced minor impacts on soil fungal community. Cultivation experiments further indicated that bacterial community from acidified soils showed a declined suppression towards pathogen growth, thereby failing in protecting host from pathogen invasion (Figure 3). Overall, our findings advance our understanding of the significance of soil health on agriculture production exerted via the microbial community assembly, and will serve to guide future soil microbiome manipulation for promotion of soil health and sustainable agriculture.
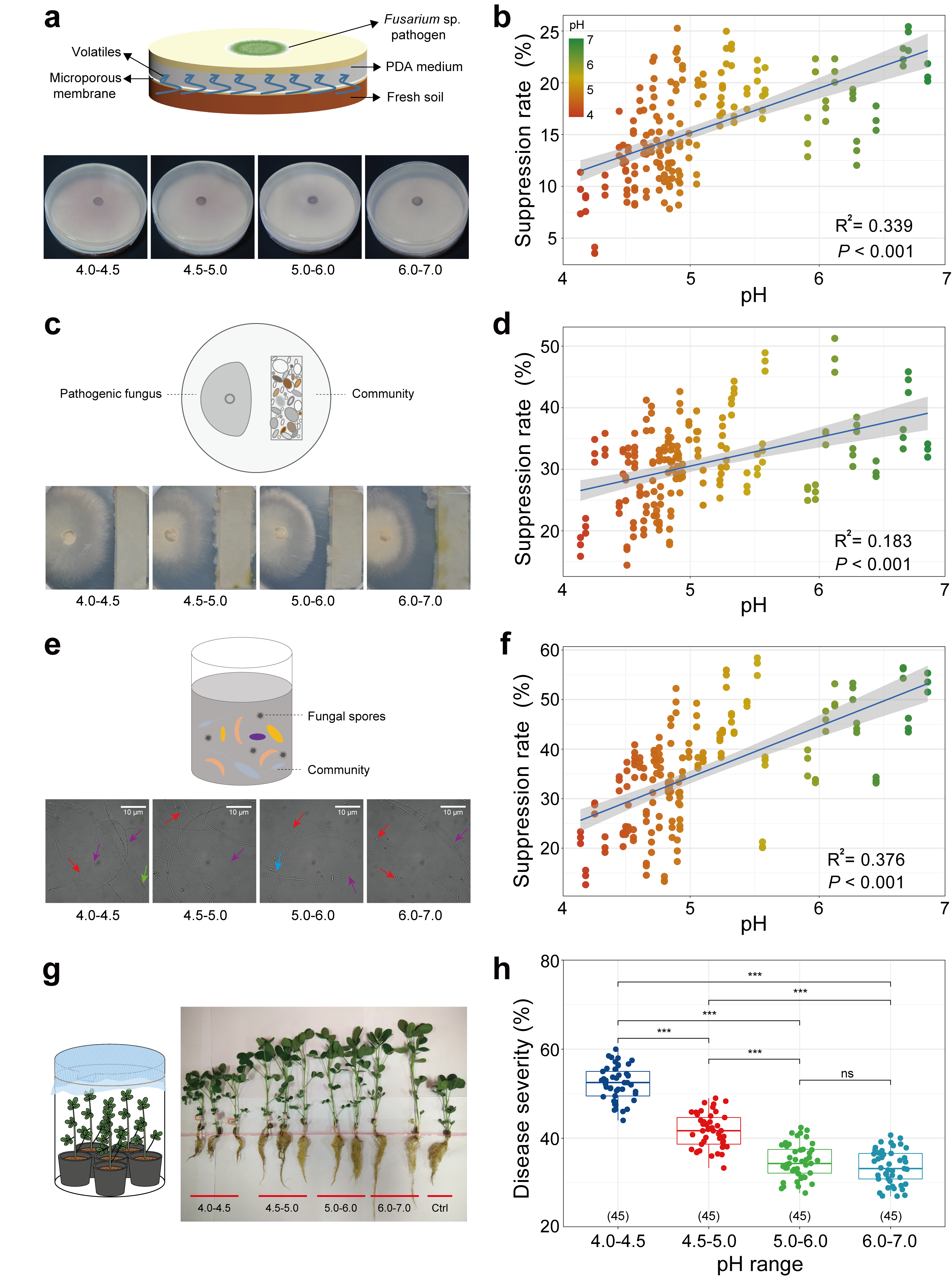 Figure 3. (a, b) Experimental set-up and results of response of mycelial growth of pathogenic fungus Fusarium sp. to soil volatiles. (c, d) Model to explore responses of the pathogenic fungus Fusarium sp. growth to soil bacterium suspension. (e, f) Experimental set-up and results of response of spore germination rate of Fusarium sp. (g) Experimental set-up and results of response of the protective efficacy of the inoculated bacterial suspension in the rhizosphere to intrusion of Fusarium sp. (h) Disease severity of peanut root after colonization of bacterial suspensions obtained from soil of different pH.
Figure 3. (a, b) Experimental set-up and results of response of mycelial growth of pathogenic fungus Fusarium sp. to soil volatiles. (c, d) Model to explore responses of the pathogenic fungus Fusarium sp. growth to soil bacterium suspension. (e, f) Experimental set-up and results of response of spore germination rate of Fusarium sp. (g) Experimental set-up and results of response of the protective efficacy of the inoculated bacterial suspension in the rhizosphere to intrusion of Fusarium sp. (h) Disease severity of peanut root after colonization of bacterial suspensions obtained from soil of different pH.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in