Adversarial domain translation networks for integrating large-scale atlas-level single-cell datasets
Published in Bioengineering & Biotechnology
Over recent years, single-cell genomics technologies have greatly improved in their capability and throughput, and are now being used to characterize a wide range of cell types through measurements of diverse data types. As such, the number and scale of such single-cell datasets has also dramatically increased. To make better use of such rich resources for new biological discoveries, there is a need for integration tools that can effectively and efficiently harmonize multi-omic datasets with even millions of cells.
Motivated by this challenge, we first explored many existing popular integration tools, and found that most of them have two limiting factors: First, their algorithms become less efficient when dealing with millions of cells. Second, most integration methods were developed to remove unwanted variation mainly caused by technical artefacts. They showed less satisfactory flexibility when handling cross-omic datasets. We then focused on how to address these two challenges simultaneously.
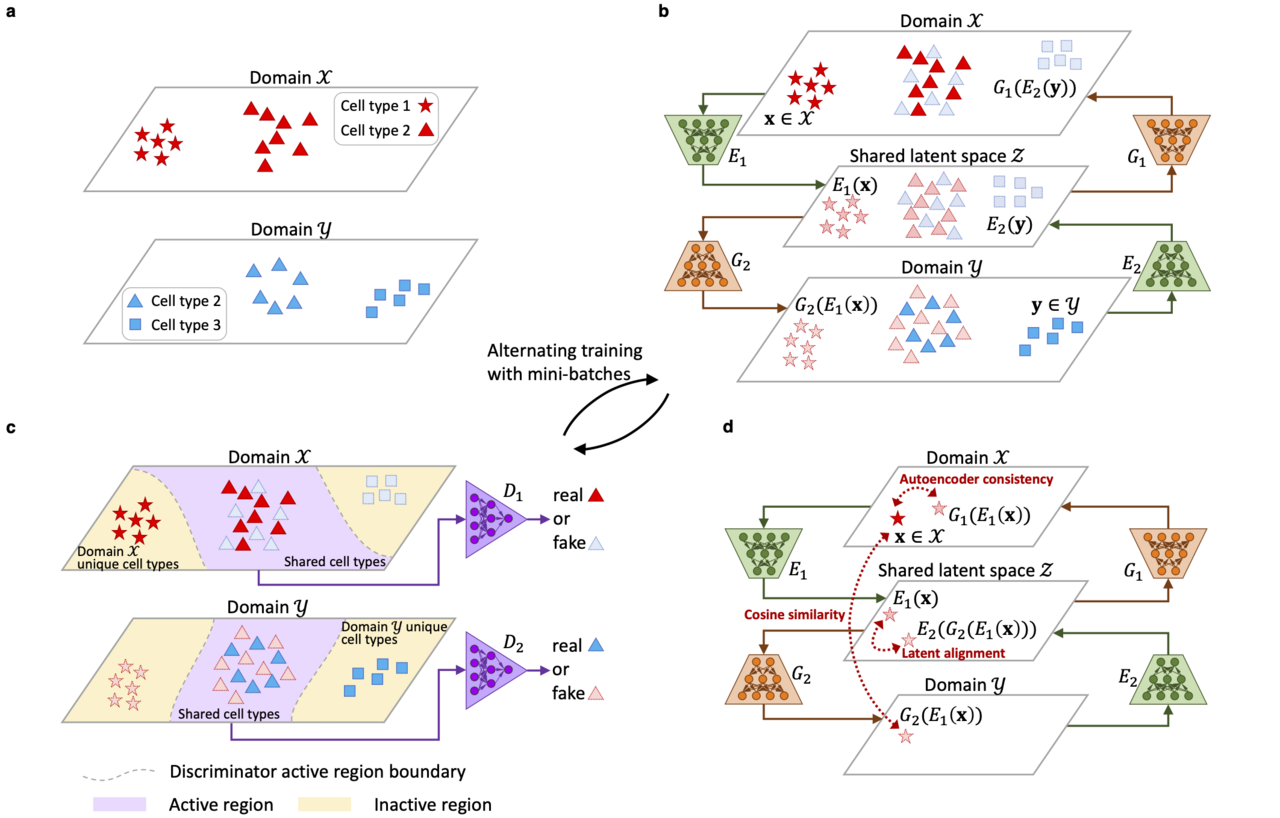
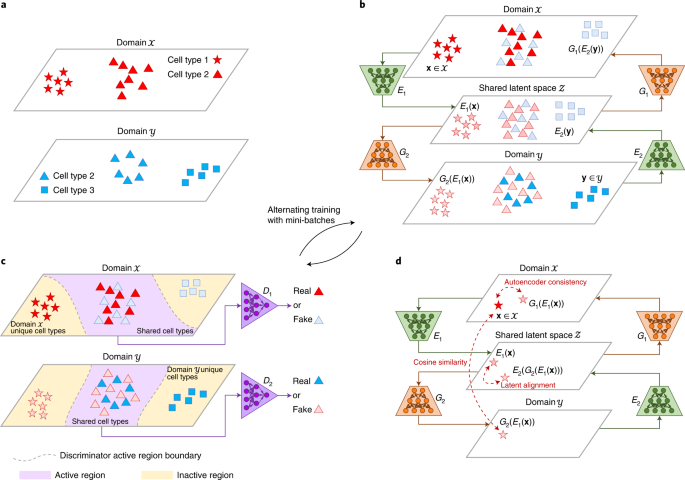
For handling large datasets, we discovered that it may not be a good idea to handle the entire dataset at once, because the computational complexity grows rapidly when the sample size gets larger. Therefore, we decided to adopt a deep generative model to turn the “curse” of large sample sizes into a “blessing”. A large sample size can help deep generative models to accurately fit the data distributions, while not incurring heavy computational costs, as the entire dataset can be decomposed into mini-batches in the training stage. To flexibly account for general unwanted variation, we formulated the integration problem as a domain translation task. Different datasets can be regarded as different domains due to the presence of domain-specific effects, which can flexibly represent unwanted variation. Our proposed method, named “Portal”, translates samples across different domains and into a shared latent space where domain-specific effects do not exist. Portal is trained utilizing the generative adversarial network (GAN) framework, which does not specify an explicit form of sample distribution, and thus has high flexibility. Being different from traditional adversarial learning, Portal is equipped with uniquely designed discriminators such that domain-specific cell types can be preserved during integration, and three regularizers to learn a robust cross-domain correspondence.
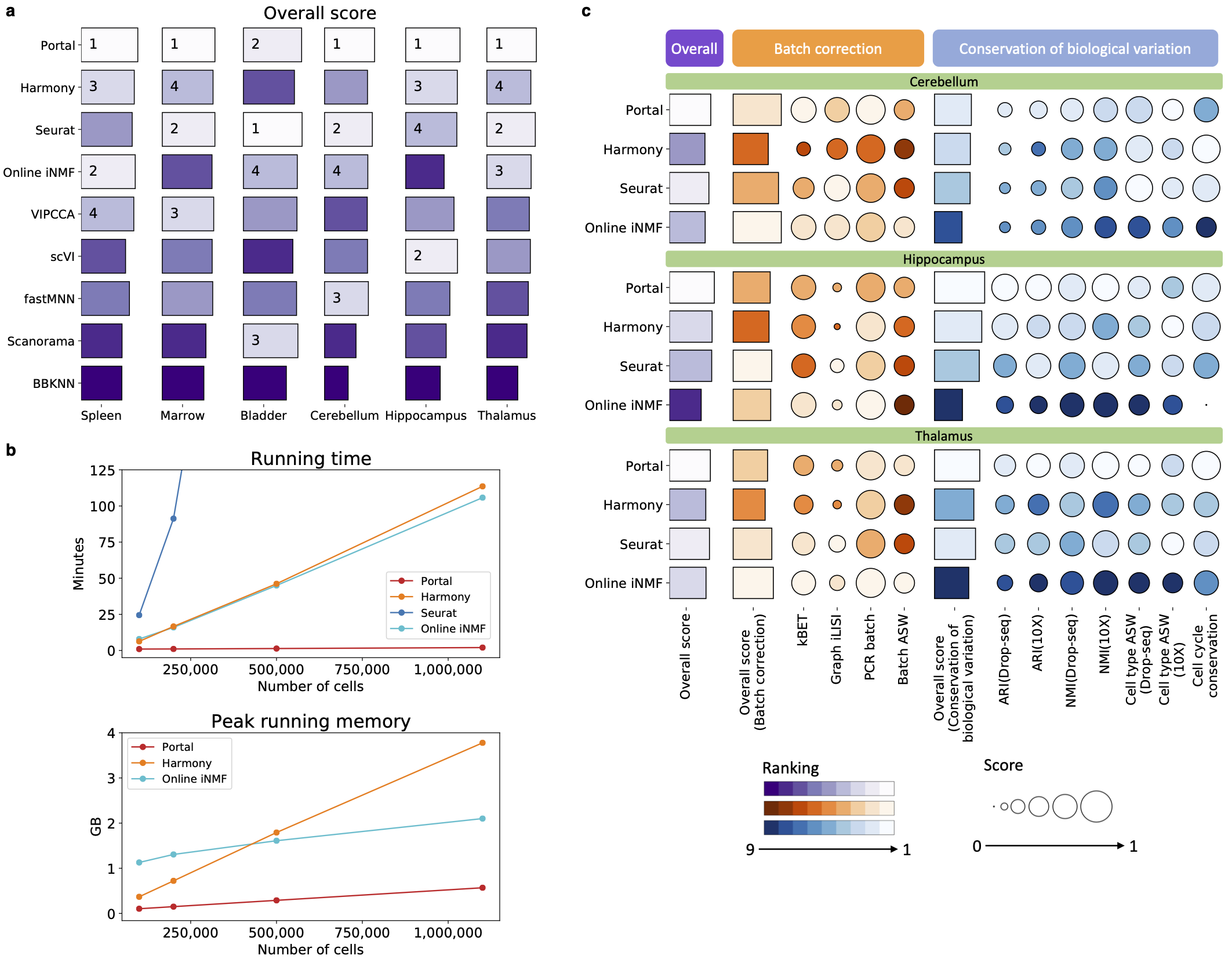
Figure 1: We benchmarked Portal and existing representative integration methods. a, The overall score jointly accounts for batch correction and biological variation conservation performance. The ranking is visualized by color gradient, where lighter color indicates better performance. b, The computational time and memory recorded using datasets with sample sizes ranging from 100,000 cells to 1,100,167 cells. c, Batch correction and biological variation conservation performance. The metrics in the category of batch correction measure how well the datasets are mixed. The metrics in the conservation of biological variation category evaluate how well biological signals are preserved.
Applying Portal to a wide range of integration tasks, we found that our approach has many advantages. First, Portal is faster and more memory efficient than other methods, including existing deep-learning based methods. It can integrate data from one million cells in about two minutes after basic data preprocessing. Second, it is highly accurate in preserving rare cell types and identifying rare subpopulations. Last but not the least, it can flexibly integrate multi-omic datasets and align datasets across multiple species. As the need for assembling comprehensive atlases and performing translational studies among diverse datasets grows rapidly, we believe that scalable and flexible integration methods such as Portal will become essential tools in life sciences research. We are excited about the innovations in single-cell technologies and generation of more atlas-level datasets that will reveal exciting new biological insights.
Full text is available at https://www.nature.com/articles/s43588-022-00251-y.
Portal software is available at https://github.com/YangLabHKUST/Portal.
Follow the Topic
-
Nature Computational Science

A multidisciplinary journal that focuses on the development and use of computational techniques and mathematical models, as well as their application to address complex problems across a range of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Physics-Informed Machine Learning
Publishing Model: Hybrid
Deadline: May 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in