After the Paper: Novel cytochrome b-containing methanogens discovered in mangrove wetlands
Published in Microbiology

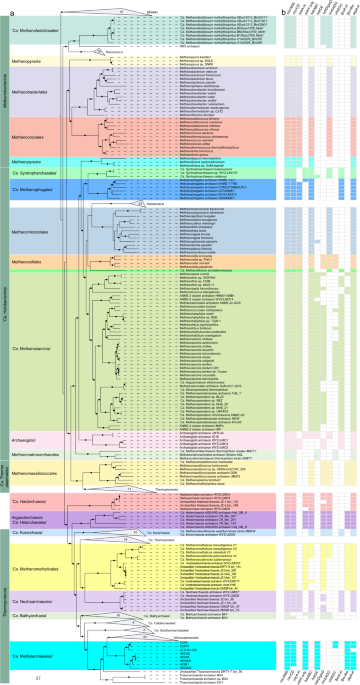
Wetland is an important source and sink of methane. Methane produced by methanogenic archaea has a great influence on Earth’s climate. Methanogenic archaea can be divided into two subgroups based on whether they use b-type cytochromes to conserve energy. Methanogens with b-type cytochromes have a wider range of substrates and higher growth yields than those without b-type cytochromes [1,2]. Here, we report seven metagenome-assembled genomes harboring methyl-coenzyme M reductase genes recovered from in mangrove nature reserves of Techeng Island and Dongzhai Harbour in China as well as Lake Towuti in Indonesia (Fig. 1a), representing a novel archaeal order, designated the ‘Ca. Methylarchaeales’ (Fig. 1b), of the phylum Thermoproteota.

Fig. 1. Sampling sites (a) and cartoon of ‘Ca. Methylarchaeales’ (b).
Metabolic analysis revealed that the ‘Ca. Methylarchaeales’ possess the complete mcrABG operon and the membrane-bound hdrDE operon (containing b-type cytochromes). Furthermore, the mcrABG operon and the hdrDE operon are located in the same contig/scaffold and are close to each other (Fig. 2a). The b-type cytochromes in the HdrDE complex of the ‘Ca. Methylarchaeales’ are integral membrane proteins with five transmembrane helical segments that harbor a nitrate reductase gamma subunit domain (PF02665), similar to the HdrE of Methanosarcina barkeri [3]. These findings strongly indicate that members of the ‘Ca. Methylarchaeales’ are b-type cytochrome-containing methanogens that use the HdrDE complex to reduce the heterodisulfide CoM-S-S-CoB of Coenzymes M and B generated in the final step of methanogenesis.
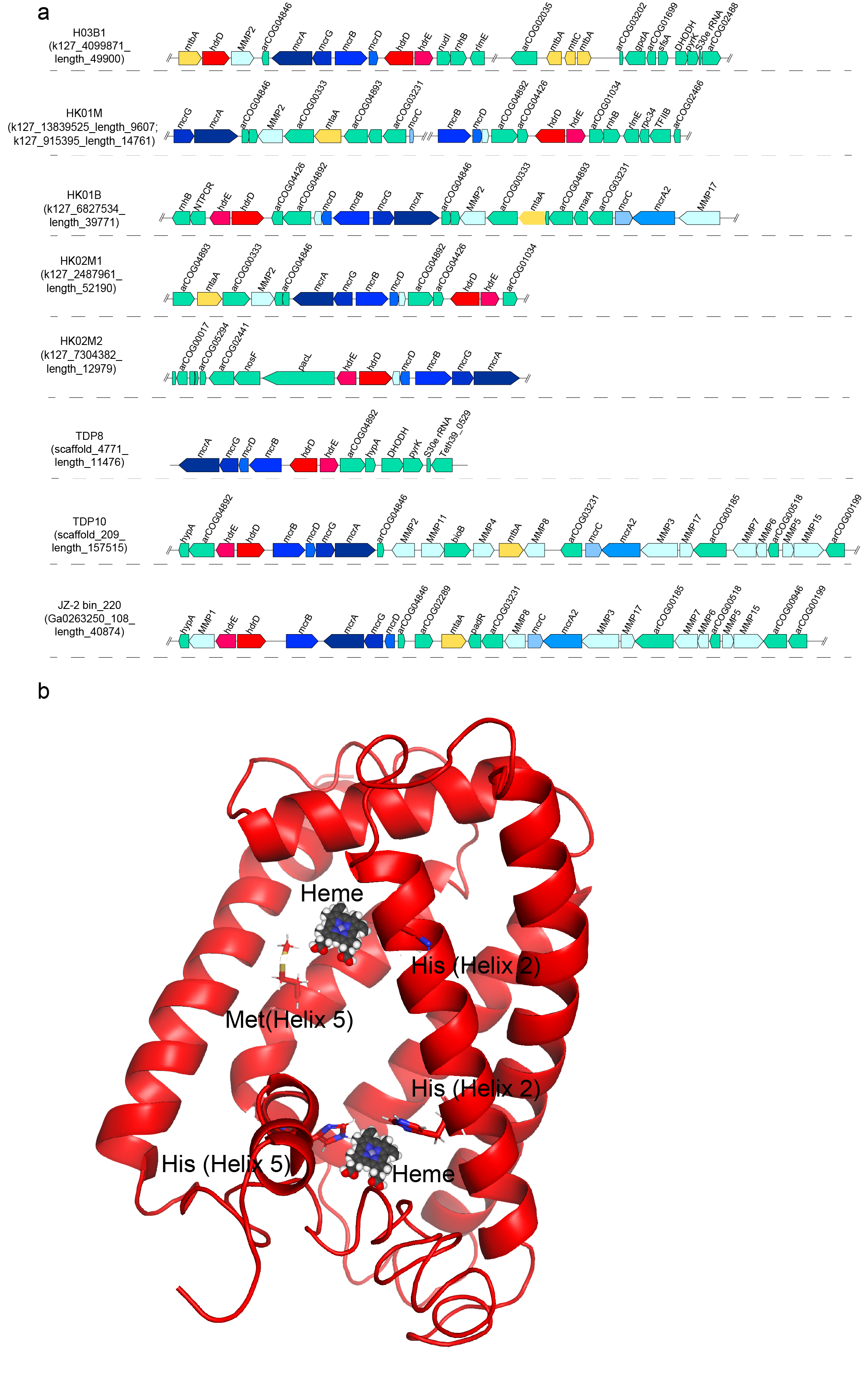
Fig. 2. Gene composition of contigs/scaffolds containing the gene cluster of mcrABG and hdrDE (a) and structural model of b-type cytochromes in HdrDE (b) in the ‘Ca. Methylarchaeales’.
Phylogenetic analyses of mcrABG genes and methanogenesis marker proteins (m4-m9) reveal that these genes in Thermoproteota (including the ‘Ca. Methanomethylicales’, ‘Ca. Korarchaeia’, ‘Ca. Nezhaarchaeales’, and ‘Ca. Methylarchaeales’) may be inherited from their last common ancestor. The wide distribution of mcr genes in archaea and their concordance with genome-based trees of many lineages in the Euryarchaeota superphylum and Thermoproteota suggest that these genes likely have originated before the divergence of the Euryarchaeota superphylum and Thermoproteota. The amalgamated likelihood estimation (ALE) analysis [4] also supports this inference that methane metabolism may have evolved relatively early in Thermoproteota. Phylogenetic analysis of NarI domain-containing cytochromes in archaea reveals that these cytochromes were already present in the last common ancestors of Thermoproteota or ‘Ca. Halobacteriota’.
By screening the Sequence Read Archive (SRA) database using the 16S rRNA and mcrA genes of ‘Ca. Methylarchaeales’ as queries, we indicate that ‘Ca. Methylarchaeales’ are widely distributed in anoxic environments such as marine sediments, freshwater sediments, deep-sea hydrothermal vents, petroleum seep, hot springs, mangrove wetlands, palm oil mill effluent, and rice paddy soils.
A previous study using molecular clock analyses to indicate that the diversification of Thermoproteota likely occurred in the early Archean Eon [5]. Archean oceans are thought to have been anoxic and contain abundant ferrous iron from hydrothermal volcanics [6,7], which would have provided sufficient raw materials for heme synthesis by methanogens. In addition, CO2, H2, and organic compounds produced by volcanic activity are transported to the early oceans [8], which provides adequate carbon and energy sources for methanogenic growth. Compared to hydrogenotrophic methanogens using electron bifurcation, methanogens using the membrane-bound electron chain have a higher energy production efficiency and growth yield, providing an advantage for members of the ‘Ca. Methylarchaeales’ described here.
References
- Mand TD, Metcalf WW. Energy conservation and hydrogenase function in methanogenic archaea, in particular the genus Methanosarcina. Microbiol Mol Biol Rev. 2019;83:e00020-00019.
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579-591.
- Künkel A, Vaupel M, Heim S, Thauer RK, Hedderich R. Heterodisulfide reductase from methanol‐grown cells of Methanosarcina barkeri is not a flavoenzyme. Eur J Biochem. 1997;244:226-234.
- Szöllősi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V. Efficient exploration of the space of reconciled gene trees. Syst Biol. 2013;62:901-912.
- Wang Y, Wegener G, Williams TA, Xie R, Hou J, Tian C, et al. A methylotrophic origin of methanogenesis and early divergence of anaerobic multicarbon alkane metabolism. Sci Adv. 2021;7:eabj1453.
- Wiechert UH. Earth's early atmosphere. Science. 2002;298:2341-2342.
- Canfield DE. The early history of atmospheric oxygen: homage to Robert M. Garrels. Annu Rev Earth Planet Sci. 2005;33:1-36.
- Canfield DE, Rosing MT, Bjerrum C. Early anaerobic metabolisms. Philos Trans R Soc, B. 2006;361:1819-1836.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in