AI-driven monitoring of concrete crack widths: a dataset for training deep learning models
Published in Materials, Research Data, and Civil Engineering
Context of research
In civil engineering, monitoring the structural health of infrastructure such as bridges, tunnels, and road surfaces is crucial. An important aspect of this monitoring is the measurement of crack widths over time.
Concrete self-healing, or the inherent ability of concrete to autonomously close its own cracks, introduces opportunities for enhancing structural durability. It is an emerging and rapidly growing field, involving multitude of laboratory and field experiments. The primary methodological challenge remains accurately assessing crack widths over time for monitoring self-healing process. Our goal was to develop a semi-automated method to accurately track the progression of self-healing at multiple fixed locations. We use image processing and a deep convolutional neural network (DCNN), which extract features from images and enable the assessment of crack widths. This procedure allows monitoring of self-healing over its stages, which is assessing the progress of recovery of the original integrity of the structure lost during cracking development.
We share a methodology and a large dataset for training deep learning models to assess concrete crack widths, particularly suited for self-healing monitoring. These resources can support the development of research techniques, ultimately contributing to improved durability and safety of concrete structures.
Crack width assessment and monitoring self-healing progress
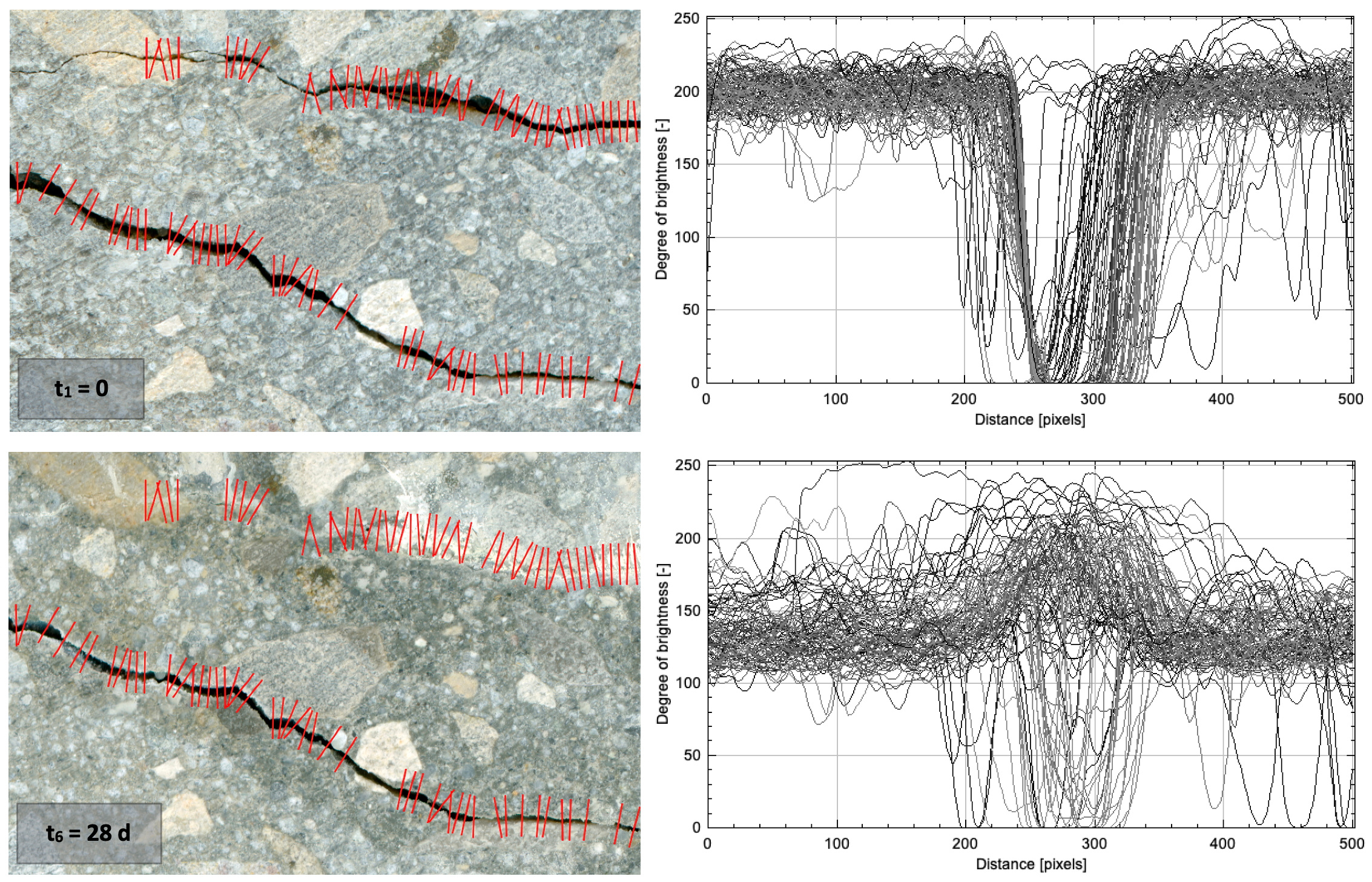
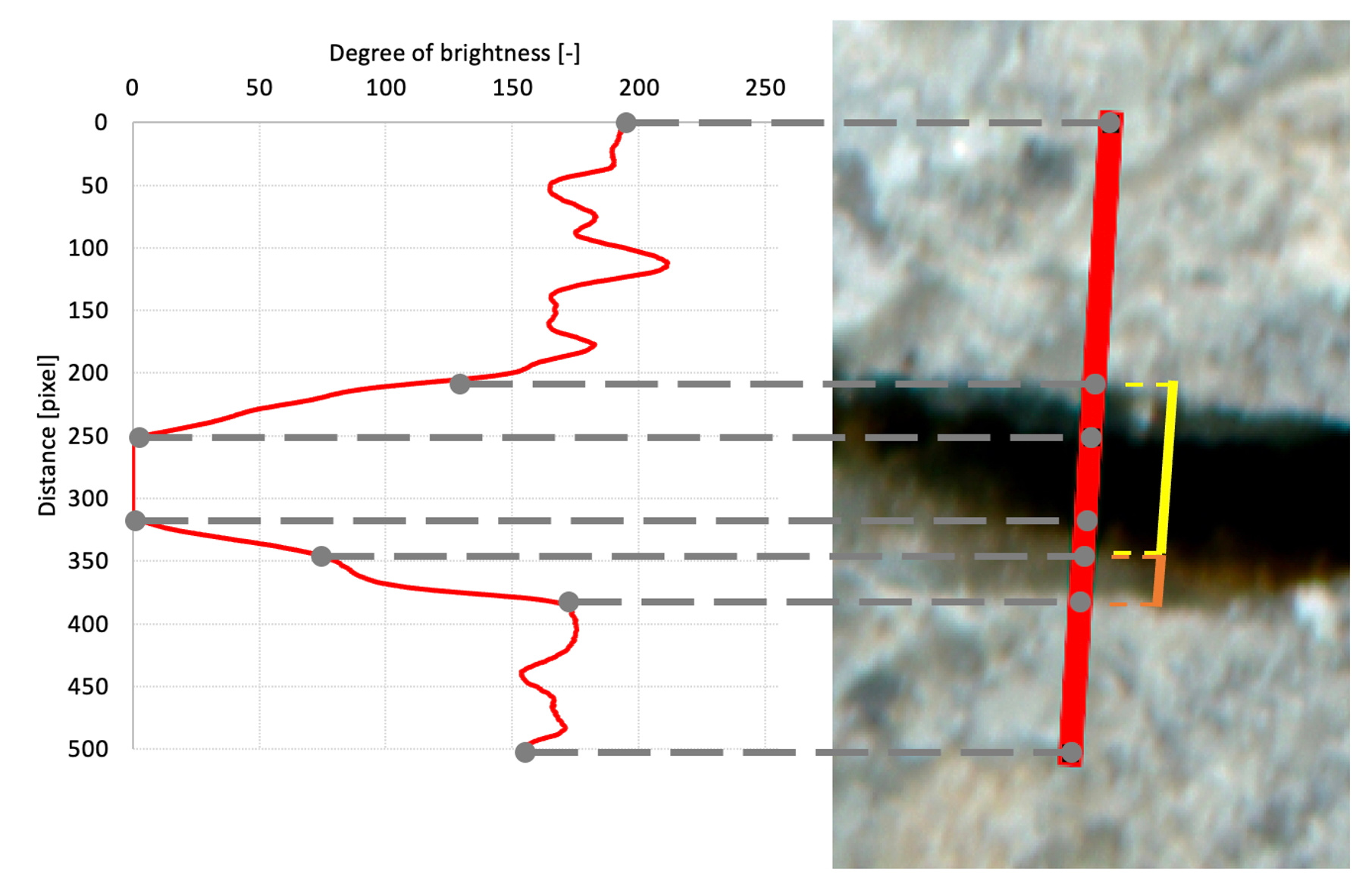
One of the advantages of our method is the consistent observation of identical locations across multiple sequential images. Achieving such consistency can be particularly difficult due to evolving cracks, shifting environmental conditions, and variable positioning of specimens relative to the imaging devices. Nevertheless, our methodology successfully addresses these issues. The method involves repeated high-resolution scanning of concrete specimens, followed by scale-invariant image registration (SIFT) and detailed brightness profile analysis along fixed gridlines intersecting cracks. This rigorous approach ensures highly accurate and consistent tracking of changes in crack geometry.
The strength of our approach lies in its systematic repeated-measures strategy for data acquisition. Each measurement is consistently performed at the same spatial locations during each phase of self-healing. This dependent data sampling significantly enhances precision of feature estimates and statistical power of tests, enabling researchers to detect subtle trends, factor effects and identify impacting factors that could be missed using traditional or independent sampling methods. High resolution and temporal continuity further support precise evaluations of factors affecting self-healing effectiveness, such as concrete age, moisture content, crack geometry, and environmental conditions.
Although developed specifically for self-healing research in concrete, our approach is also applicable to broader contexts where accurate crack width measurements are needed, such as general structural health monitoring or artificial intelligence-based damage assessments.
What is in the dataset
Our dataset was collected using high-strength concrete specimens, carefully prepared, aged, and cracked to create conditions for observing self-healing. High-resolution scans were employed to capture detailed images of specimen surfaces at various healing stages. The resulting dataset contains 19,098 records, each with comprehensive brightness profiles collected along gridlines intersecting visible cracks. The records include operator crack width measurements, serving as reference values, along with benchmark measurements provided by a convolutional neural network and analytical algorithm.
Key components of the dataset include:
- Brightness profiles along each intersecting gridline.
- Manual reference measurements.
- Predictions from a deep convolutional neural network model and analytical edge detector.
- A deep convolutional neural network model trained for our own research.
- High-resolution scans of concrete surfaces at multiple stages of healing.
- Consistent, fixed gridlines intersecting crack paths.
This large and comprehensive dataset is well-suited for training and evaluating deep learning models aimed at crack width estimation.
Sharing dataset
Published researches demonstrate the considerable potential of deep learning methodologies in monitoring crack widths. Providing open access to a detailed, validated dataset significantly reduces the required resources and time for individual research groups, encouraging innovation and enhancing research efficiency. By sharing our dataset, we intend to encourage and enable diverse research projects.
We invite researchers from various fields, including structural engineering, materials science and computational intelligence, to utilize this data to:
- Develop and refine convolutional neural networks for accurate crack width estimation.
- Use your own or our pre-trained model to investigate self-healing dynamics under diverse conditions.
- Integrate precise crack width monitoring into broader structural health monitoring and predictive maintenance systems.
- Extend the methodology and explore new applications in civil engineering, materials engineering, predictive maintenance, or other relevant fields.
For detailed methodology, dataset access, and further information, please refer to our publications:
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in