An Antarctic plasmid goes viral
Published in Microbiology

The paper in Nature Microbiology is here: http://go.nature.com/2iMIyll

Antarctica leaves everyone with a sense of awe – its beauty, wildness and remoteness, setting it apart from anywhere else. It is the planet’s most important continent for influencing global climate and ocean ecosystem function with its massive reserve of ice buffering against the effects of global warming.

Antarctica has a deserved reputation for being untamed and unforgiving, thereby, somewhat ironically, luring scientist and artist expeditioners alike with the tantalizing prospect of experiencing unique wild life and ever-changing sculpted icescapes.


Beyond Antarctica’s visual splendour lies a whole other level of wonder – the microbial world – the unseen majority that controls ecosystem function. Globally, environmental microorganisms literally make the world go around – not physically, but in terms of performing nutrient cycling that maintains the existence of all other life forms, and hence, the health on the planet. Antarctica’s Southern Ocean microorganisms fix a disproportionately large amount of the world’s carbon dioxide, using the Sun’s light as energy to grow into massive blooms in seawater and form dense brown communities within sea ice – notably, the Antarctic sea ice extent represents the largest physical seasonal change on the planet, growing up to 20 million km2 through winter and then largely melting away in summer. The marine food-web starts with primary producing phytoplankton and heterotrophic remineralizing microorganisms and branches out through krill to all other macroscopic marine life – sponges, fish, sea birds and sea mammals.


The Antarctic continent also harbours diverse lake systems – from subglacial behemoths like Lake Vostok and ancient wonders oozing out Blood Falls, to the plethora of diverse, marine-derived systems in the Vestfold Hills and Rauer Islands near Australia’s Davis research station. The Vestfold Hills’ lakes are each, in effect, unique time capsules that show us how marine life has evolved after being isolated from the ocean about 3-5,000 years ago. Because of the limited nutrient input, changes in oxygen levels, salinity and overall geochemistry, the biology of these systems has become progressively microbially dominated, supporting the existence of few, higher-trophic (metazoan) predators. During this remarkable evolutionary process, viruses have emerged as particularly key components of the communities, affecting community dynamics, productivity and nutrient cycling, and the flow of genes between community members.
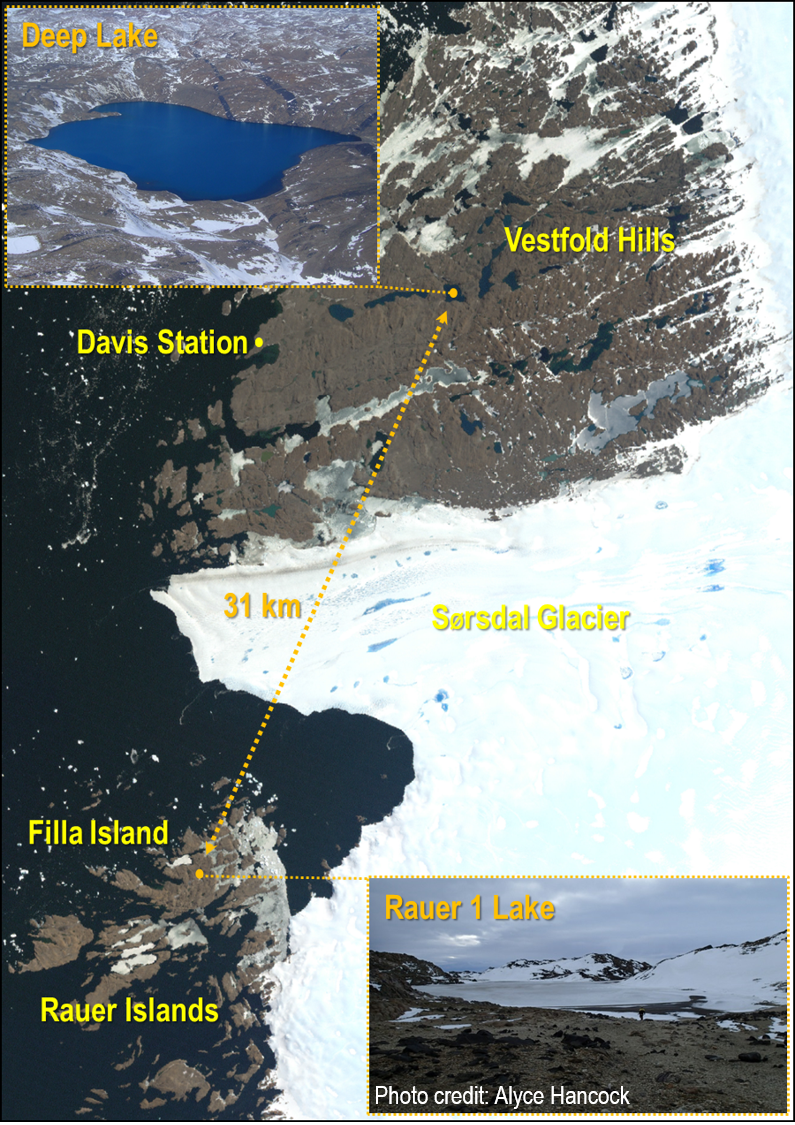
How do we know about Antarctic viruses and the roles they perform? Firstly, we can see ‘virus-like particles’ (VLPs) in water samples, under the microscope. Secondly, we can see viral sequences in metagenomics data and track their interactions by studying CRISPRs (host-defence systems), that by virtue of being primitive immune systems, provide an inventory of what the CRISPR system has defended against. Thirdly, viruses can be isolated in the laboratory and their interactions with hosts studied.
In a recent paper led by my group in Nature Microbiology, all of these factors came together to lead us on a journey that ultimately abolished the final boundary that distinguished viruses from plasmids. From previous metagenomics studies by my group of a hypersaline lake, Deep Lake, that remains liquid even when the water temperature drops to -20°C, we found that a low complexity community of haloarchaea dominate the system and are promiscuous in supporting a high level of intergenera exchange. In searching for the mechanisms of exchange, using metaproteomics we further discovered a complex network of viruses interacting with their haloarchaeal hosts.

In September 2014, Susanne Erdmann joined my group as an EMBO Fellow from Roger Garrett’s lab in Copenhagen. Building upon her expertise of studying viruses that infect hyperthermophiles (Sulfolobus), Susanne set about isolating viruses using the Antarctic haloarchaea as hosts.
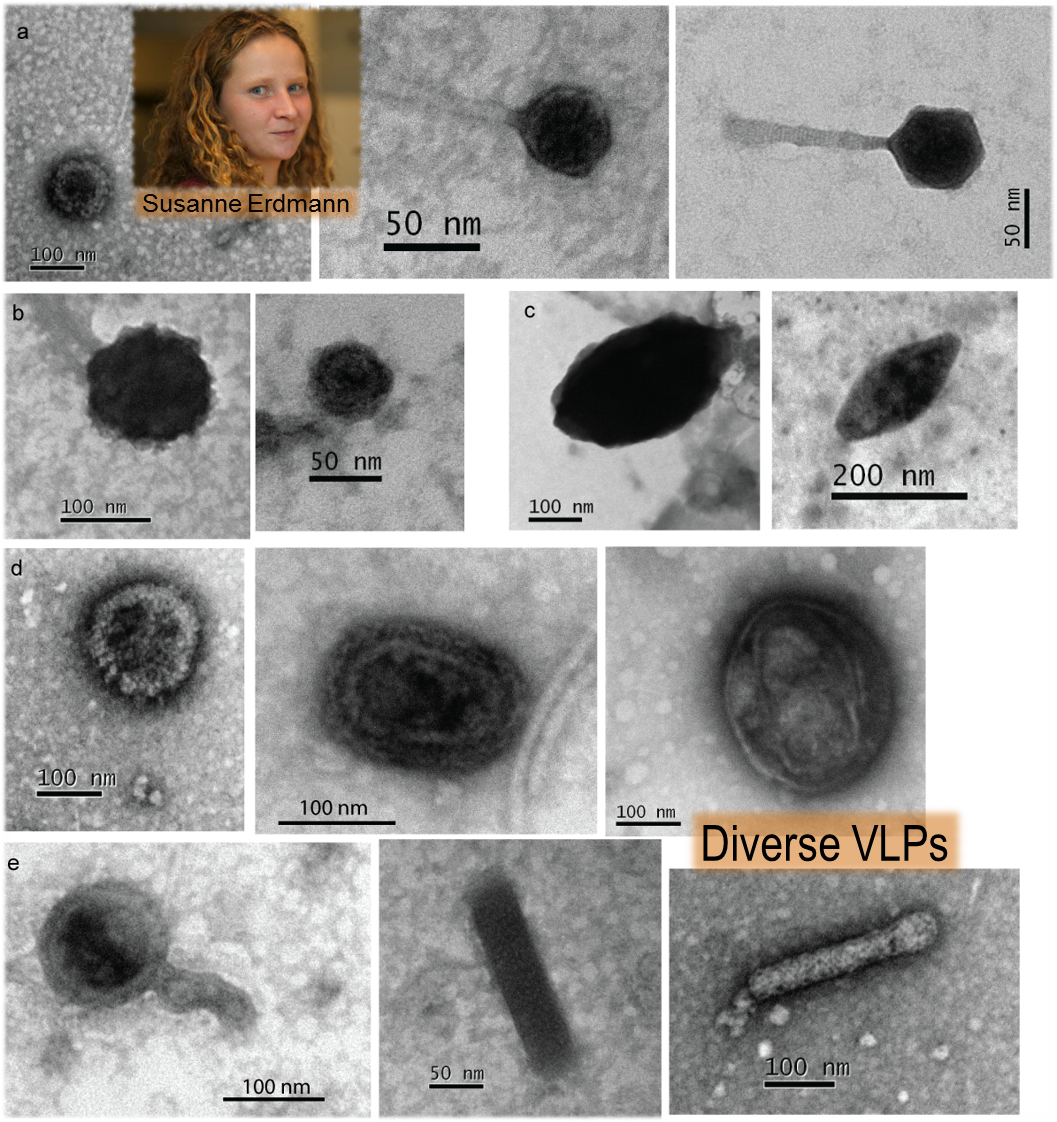
Susanne’s first step was visualizing VLPs, which she did very successfully, identifying numerous sizes and morphologies, and then sequencing their genomes. And this was where the discovery was made – a DNA sequence revealing genes typical of a plasmid and not typical of a virus. Our institution, UNSW Sydney, fortunately has excellent mass spectrometry facilities for proteomics. By working with our collaborators Mark Raftery and Ling Zhong, Susanne determined that the VLPs the plasmid was packaged in were in fact membrane vesicles that contained plasmid-derived proteins. Some of the proteins resemble those used to form the cage that surrounds eukaryotic transport vesicles (e.g. COPI), so there are nice prospects for the future to use cryoEM to study the structures of our ‘plasmid vesicles’ to find out how similar they are.
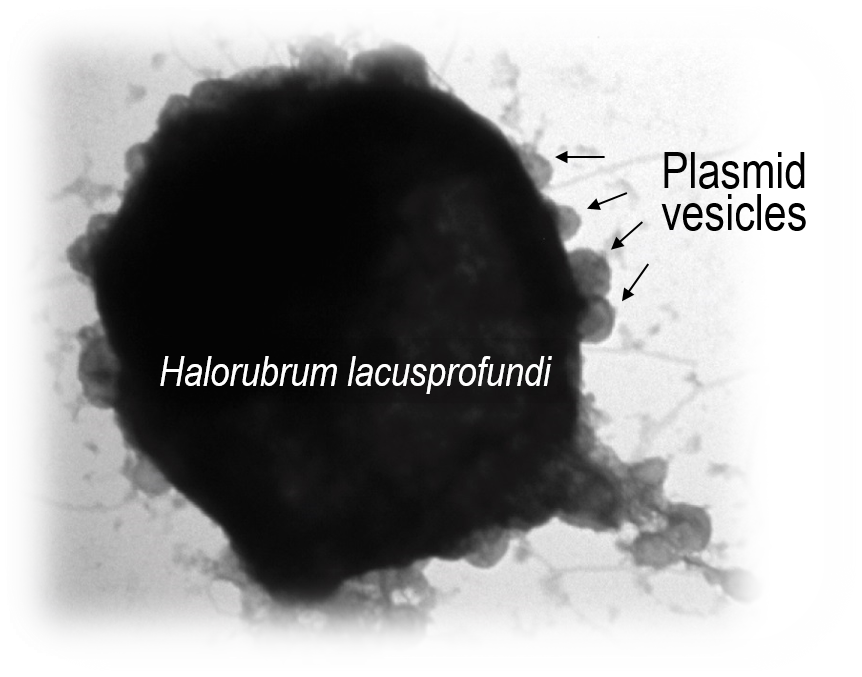
Susanne was also able to demonstrate that the plasmid integrates into the host chromosome, resolves out with host DNA, and packages the plasmid into ‘plasmid vesicles’ that then infect plasmid-less hosts. So our findings remove the last remaining major distinction between viruses and plasmids by showing that this type of extrachromosomal element that most closely resembles a plasmid (not a virus), encodes proteins that go into the particles that contain the plasmid DNA, and the particles are capable of infecting other cells.
Patrick Forterre, Mart Krupovic and others have previously speculated that host-derived membrane vesicles could have been a forerunner of virus particles, leading Forterre to even ponder about what defines a living organism. Our findings certainly work with the ‘escape hypothesis’, which denotes that viruses evolved as fragments of cellular DNA capable of infecting other cells so they could escape and propagate in a wider range of hosts. In a News and Views article, Forterre and coworkers reflect on our findings and speculate about what they mean for the evolution of viruses, including even advocating that ‘plasmid vesicles’ (a term we coined), in fact be termed ‘plasmidions’.
As a closing reflection on viruses and what they are, I offer a short story that I often tell undergraduate students that arose from a discussion I had with a cosmologist colleague, Charley Lineweaver. Charley is very interested in biology and evolution on Earth and in the Universe and asked me whether I considered a virus a life form – something that is alive. With an air of knowing, I said, ‘well Charley, the prevailing view is that life needs to be autonomous, and as a virus needs a host to propagate, by this definition, it would not be considered alive.’ To this Charley replied, ‘Well what about humans’. I looked at him quizzically and said, ‘What do you mean, humans?’ To which he responded, ‘Don’t they need a planet?’ I’ve always remembered this as a nice example of perspective, the value of interdisciplinary discussion, and also a reminder to be a little humbled about one’s ability to comprehend the world, let alone the Universe.
Cavicchioli, R. Microbial ecology of Antarctic aquatic systems. Nat. Rev. Microbiol. 13, 691–706 (2015).
Forterre, P. To be or not to be alive: How recent discoveries challenge the traditional definitions of viruses and life. Stud. Hist. Philos. Biol. Biomed. Sci. (2016). http://dx.doi.org/10.1016/j.sh...
Forterre, P. & Krupovic, M. in Viruses: Essential Agents of Life (Ed. Witzany, G.) 43–60 (Springer, Dordrecht, 2012). ISBN 978-94-007-4899-6
Kelly, J. Beyond Woop Woop – John Kelly in Antarctica. (What Gina Did Next, Melbourne, 2015). ISBN 978-0-9943184-0-4
Soler, N., Krupovic, M., Marguet, E. & Forterre, P. Membrane vesicles in natural environments: a major challenge in viral ecology. ISME J. 9, 793–796 (2015).

Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in