
Bacteria can sense their population density through a process called quorum sensing (QS). QS relies on the secretion, detection and response to extracellular signaling molecules termed autoinducers (AI). Thereby, QS controls various complex behaviors in Vibrio cholerae, including virulence factor expression and biofilm formation (1,2). At the core of the QS pathway, four homologous redundant small RNAs (Qrr1-4, sRNAs) regulate the expression of multiple target genes in a cell-density dependent manner and thereby orchestrate collective behavior (3).
In our study, we performed RIL-seq (RNA interaction by ligation and sequencing) (4,5) to identify RNA-RNA duplexes bound by the RNA chaperone Hfq in V. cholerae. Thereby we identified hundreds of previously unknown Hfq-dependent RNA-RNA interactions at low and high cell density respectively. To share our global map of interactions we created a dynamic and searchable web interface, available at http://rnaseqtools.vmguest.uni-jena.de/
The interaction network revealed sRNA-mRNA interactions as well as RNA duplexes involving two sRNA regulators and among the latter, one specific interaction caught our interest. We discovered that a previously uncharacterized sRNA formed RNA duplexes with all four Qrr sRNAs but no other RNA in our RIL-seq experiment. Therefore, we named this sRNA QrrX. sRNAs that base-pair with and inhibit the activity of other sRNAs are called sponge RNAs (6).
We monitored RNA levels of QrrX and Qrr1-4 over various stages of growth, revealed extensive RNA-duplex formation between QrrX and each of the four Qrr sRNAs and demonstrated that pulse induction of QrrX led to drastic reduction in Qrr1-4 stability. However, a single nucleotide exchange (SNE) in the base-pairing region of QrrX almost fully restored Qrr1-4 stability and we could restore the observed phenotype by introduction of the compensatory mutation.
How can base-pairing affect RNA stability? To address this question, we monitored Qrr4 RNA levels by Northern blot analysis in four different strains: wild-type, ∆rng, rneTS and ∆rng/ rneTS. RNase E and RNase G are two partially redundant endoribonucleases which have been previously described to be involved in sRNA-mediated mRNA degradation (7). Lack of RNase E alone or in combination with RNase G lead to elevated RNA levels of QrrX and Qrr4. We measured Qrr4 RNA levels after pulse induction of QrrX and addition of rifampicin: Lack of RNase G had only a slight effect on Qrr4 stability whereas lack of RNase E resulted in increased stability. Hence, we concluded that RNase E is the major ribonuclease involved in Qrr4 turnover.
Further, we were interested in the factors that control qrrX transcription. Therefore, we performed a genetic screen using a plasmid-based library of the V. cholerae genome and a qrrX::lacZ reporter. A screen for blue colonies revealed activation of qrrX expression by the uncharacterized LysR-type transcriptional regulator Vca0830, which we named QrrT. In further experiments, we could validate binding of QrrT to the qrrX promotor.
QS is dependent on post-transcriptional gene regulation by the Qrr1-4 sRNAs. These sRNAs control two major transcription factors in V. cholerae, AphA and HapR, which are key regulators in collective behaviors like biofilm formation. Therefore, we speculate that regulation of Qrr1-4 sRNAs by QrrX might also affect AphA and HapR protein levels and by quantitative Western Blotting, we discovered that lack of qrrX resulted in increased AphA and decreased HapR protein levels.
Cell-density dependent bioluminescence can be used as a read-out for QS performance. Our wild-type strain lacks the ability to produce light, however, introduction of the luxCDABE operon from V. harveyi into V. cholerae results in cell-density dependent light production2. In contrast to our wild-type strain, which quickly reached maximal light production, lack of qrrX resulted in delayed and overall reduced bioluminescence.
Finally, we measured biofilm formation of wild-type and ∆qrrX strains in microfluidic chambers using confocal microscopy. Indeed, biofilm thickness of cells deficient for QrrX was significantly increased compared to wild-type cells. In a competition assay, when wild-type and ∆qrrX were seeded at an initial ratio of 1:1, cells lacking the qrrX gene had a competitive advantage against wild-type V. cholerae cells.
To summarize, our study identified and characterized an RNA sponge regulating QS-controlled phenotypes such as bioluminescence and biofilm formation by direct base-pairing to and thereby inhibiting the Qrr1-4 sRNAs (Fig. 1).
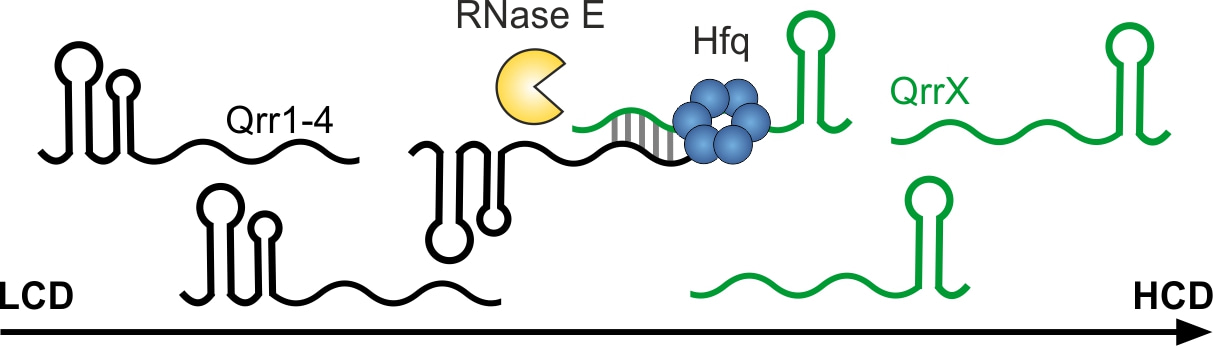
Fig. 1: QrrX binds to the Qrr1-4 sRNAs and drastically reduces their stability.
The full paper (“An RNA sponge control quorum sensing dynamics and biofilm formation in Vibrio cholerae”) is available at Nature Communications: https://www.nature.com/articles/s41467-022-35261-x
- Papenfort, K. & Bassler, B. L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol.14, 576–588 (2016).
- Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K. & Bassler, B. L. Parallel Quorum Sensing Systems Converge to Regulate Virulence in Vibrio cholerae. Cell110, 303–314 (2002).
- Shao, Y., Feng, L., Rutherford, S. T., Papenfort, K. & Bassler, B. L. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J.32, 2158–2171 (2013).
- Melamed, S. et al. Mapping the small RNA interactome in bacteria using RIL-seq. Nat. Protoc.13, 1–33 (2018).
- Melamed, S. et al. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol. Cell63, 884–897 (2016).
- Denham, E. L. The Sponge RNAs of bacteria – How to find them and their role in regulating the post-transcriptional network. Biochim. Biophys. Acta BBA - Gene Regul. Mech.1863, 194565 (2020).
- Quendera, A. P. et al. RNA-Binding Proteins Driving the Regulatory Activity of Small Non-coding RNAs in Bacteria. Front. Mol. Biosci.7, 78 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in