Annotating the unknown: Functional Gene Annotation using Untargeted Metabolomics
Published in Microbiology

Six years ago, a consortium of research groups with expertise in genomics, structural biology and biochemistry combined forces in a NIH-NIAID-funded project called FLUTE. Their goal? To discover the function of genes of unknown function that Mycobacterium tuberculosis (Mtb) needs to grow. This blog describes one of their success stories1.
A new frontier
We live in what some people call the post-genomic era. Thousands of genomes encoding millions of genes have been sequenced, and new ones are added every day. But in the wake of this genomic data explosion, a new frontier has appeared: finding the function of all these genes. Even in the best characterized organisms, about one in three genes remains unannotated while the annotation of most others relies on bio-informatic prediction alone. This annotation gap has severely handicapped our fundamental understanding of life.
A rich source of drug targets
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is no exception. There are over a thousand Mtb genes without known function and many of those turn out to be essential for the bacteria to grow and infect humans. Though commonly thought of as a disease of the past, TB still kills about 1.5 million people each year, making it the leading cause of death by a single pathogen. Most of the clinically used TB drugs were developed in the fifties and sixties of the previous century, so drug resistance has now become widespread. As a result, there is a pressing need for new drugs against TB. Such drugs should ideally kill Mtb but leave other bacteria (like those in our gut) untouched. Drugs targeting essential genes of unknown function hold promise to do exactly that, making the pool of essential genes of unknown function a rich source of potential drug targets.
Untargeted metabolomics to discover the function of genes
The Mtb gene rv3722c ranked high on the FLUTE consortium’s list of essential but unannotated genes. Using an in vitro technique called activity-based metabolite profiling, Kyu Rhee’s group discovered that Rv3722c is the primary aspartate aminotransferase in Mtb. Aspartate aminotransferases are enzymes that transfer nitrogen from glutamate to aspartate and back. Follow-up experiments in bacteria not only showed that Rv3722c is essential for the biosynthesis of aspartate, but also revealed that the nitrogen that Rv3722c transfers to aspartate is used for the synthesis of a myriad of downstream metabolites by enzymes that have all been shown to be essential for Mtb2, as you can read in this blog by Michael Berney. Perhaps not surprising, mouse infections performed by Eric Rubin’s group at Harvard TH Chan School of Public Health showed that Rv3722c-deficient bacteria were unable to establish an infection in mice, revealing an essential role for nitrogen distribution in the pathogenicity of Mtb.
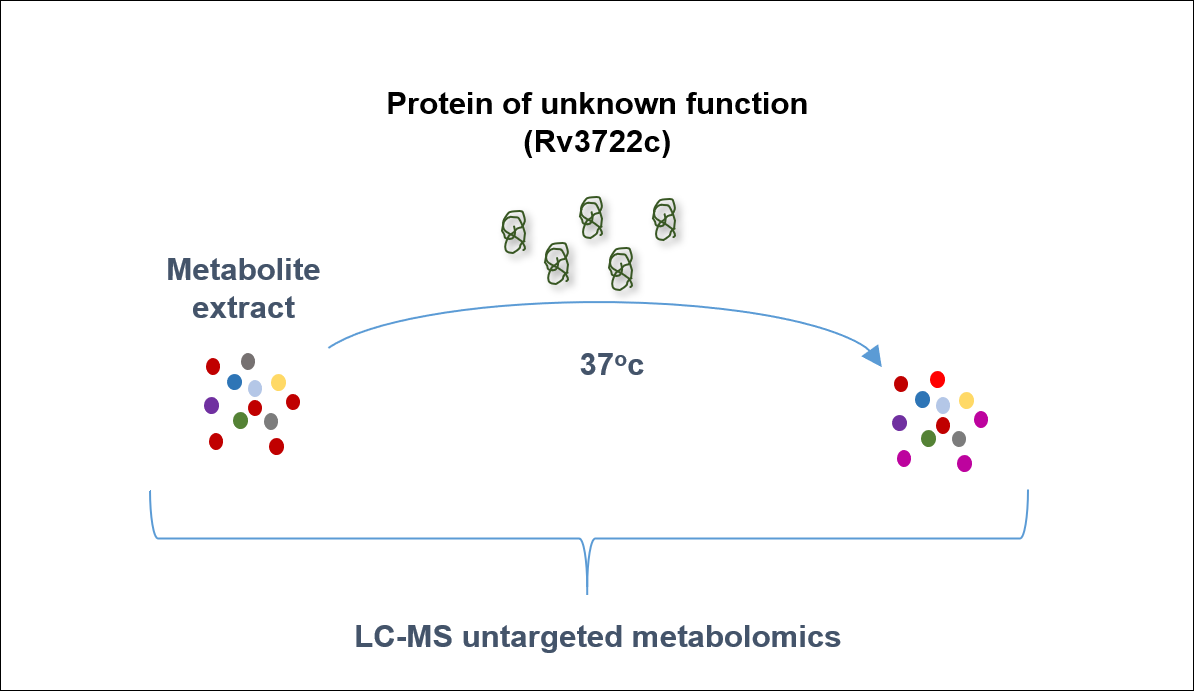
Activity-based metabolite profiling is a technique for untargeted protein annotation.
A new version of an old enzyme
Aspartate aminotransferases are among the most well-studied enzymes, so it was a surprise that our unknown was not recognized before. The reason for this was discovered when Jim Sacchettini's group at Texas A&M solved the crystal structure. Unlike the classical well-studied aspartate aminotransferases, Rv3722c contains an extended N-terminal domain of unknown function. Phylogenetic analysis showed that this domain was only present in a select group of bacteria and not in humans.
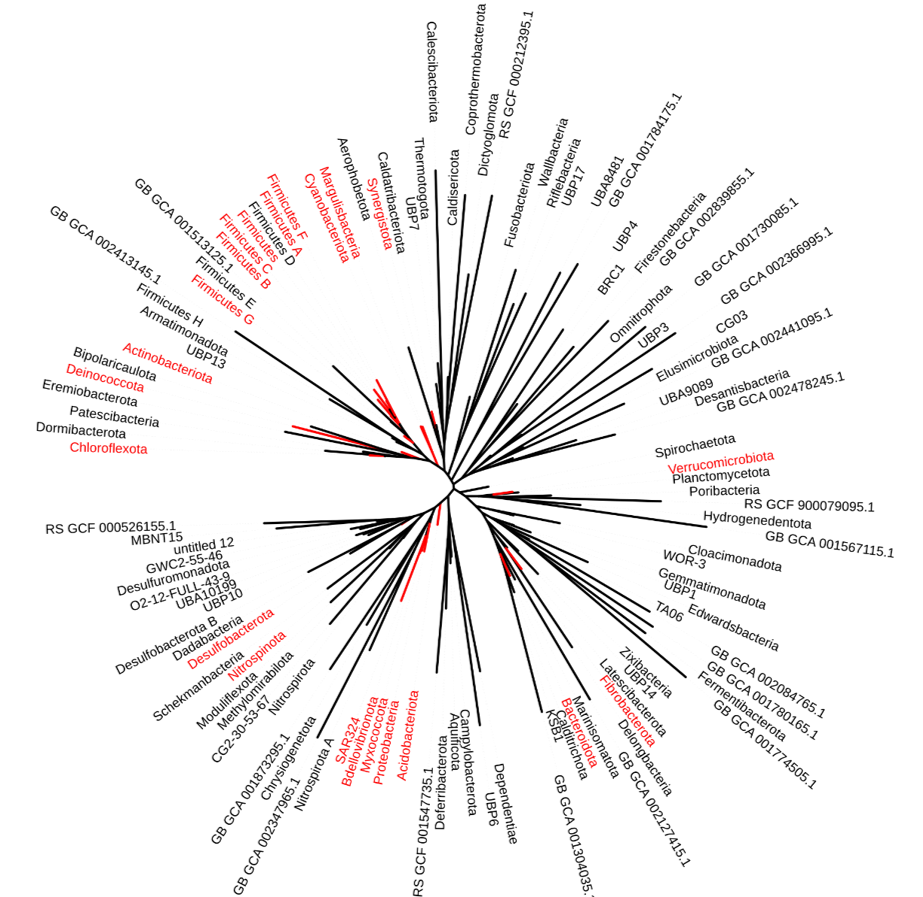
Rv3722c-homologs are only present in a select group of bacteria (highlighted in red).
How can this help to fight tuberculosis?
Aminotransferases have been hard to target specifically with small molecules. The relatively narrow species distribution of this new subclass of aspartate aminotransferases, however, opens the door for selective targeting of the first step of an essential pathway in Mtb.
References
- Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis. Jansen RS, Mandyoli L, Hughes R, Wakabayashi S, Pinkham JT, Selbach B, Guinn KM, Rubin EJ, Sacchettini JC, Rhee KY. Nat Commun. 2020.
- Derailing the aspartate pathway of Mycobacterium tuberculosis to eradicate persistent infection. Hasenoehrl EJ, Rae Sajorda D, Berney-Meyer L, Johnson S, Tufariello JM, Fuhrer T, Cook GM, Jacobs WR Jr, Berney M. Nat Commun. 2019.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in