Another Piece Added to the Structure Universe of Electron Transport Chain
Published in Cell & Molecular Biology

Mitochondrial oxidative phosphorylation (OXPHOS) system, encompassing the electron transport chain (ETC) complexes I-IV (CI-CIV) and the ATP synthase or complex V (CV), cashes out the energy stored in reduced electron carriers NADH and FADH2 into ATP. In such way the OXPHOS system is the major energy supplier for most cellular organisms using aerobic respiration at least as one of their metabolic strategies or during one stage of their life cycles. The trans-membrane complexes, usually found on the inner membrane of mitochondria (IMM), are classic ‘molecular machines’ that are large, involving profound conformational changes to carry out catalysis and drawing research attentions since the early onset of biochemistry. Nowadays with the powerful tool of Cryo-electron microscopy (Cryo-EM), structural and mechanistic studies of mammalian complexes I-V are plenty1. Meanwhile researches into the diversities of the OXPHOS system among different clades of the Eukaryota domain are also fast accumulating2,3, revealing to us a beautiful and dynamic picture of how these energy supplying molecular machines evolved during the long history of lives after the endosymbiosis event.
We chose to investigate the ETC complexes of Euglena gracilis, a free-living, flagellated protist utilizing at least aerobic respiration, anaerobic fermentation and photosynthesis via its secondary plastid, as its metabolic strategies. Past works, including our own, have already presented complexes I-V structures from eukaryotic clades Opisthokonta (fungi and metazoan)4, Archaeplastida (red/green algae and vascular plants)3 and Alveolata (ciliates, apicomplexans and so on)2,5. However, the clade Excavata, now split into three supergroups including Discoba, Metamonada and Malawimonadida, lacked high resolution ETC complex structure. E. gracilis was one of the model organisms of the supergroup Discoba characterized by the discoidal shape mitochondrial cristae. In light of our published structure of Tetrahymena thermophila ETC megacomplex which exhibited an overall shape of ~180° arc adapting perfectly to the tubular cristae of ciliates (Figure 1A) 5, we decided to have a look at the relation between E. gracilis’s ETC and its discoidal cristae (Figure 1B).
The outcome of such effort was quite rewarding. Although the overall architecture of E. gracilis’s ETC (Eg-ETC) supercomplex (SC) I+III2+IV, or respirasome, does not differ too much compared to its mammalian counterpart, closer examination reveal that Eg-CIII2 has shifted towards the peripheral arm (PA) of Eg-CI for ~41 Å (Figure 1C). Canonically, association between CI and CIII2 within SC I+III2+IV is via the distal end of CI membrane arm (MA) and the peripheral arm is not involved in such process. For Eg-SC I+III2+IV however, CIII2 associates CI PA subunit NDUFA9 and the interaction between CIII2 and the distal end of CI MA is now bridge by Eg-CIV. This in turn explains why SC I+III2, the recognizably most conserved supercomplex form of eukaryotic ETC, does not exist in E. gracilis, because losing Eg-CIV will break the CIII2-CI MA interaction in such peculiar respirasome architecture. More surprisingly, when viewed form the distal end of CI MA, a slight negative curvature is discovered between Eg-CI and the rest Eg-CIII2+CIV part. This is due to the presence of several Eg-specific subunits NDUEG7-9 wedging in between Eg-CI and CIII2 from the inter membrane space side of IMM. The only sensible explanation is that such arrangement adapts to the negative membrane curvature found in the discoidal cristae, between its positively curved rim and flat central region (Figure 1B). This is maybe the second example of ETC supercomplex adaptation to cristae morphology that is not lamellar5.
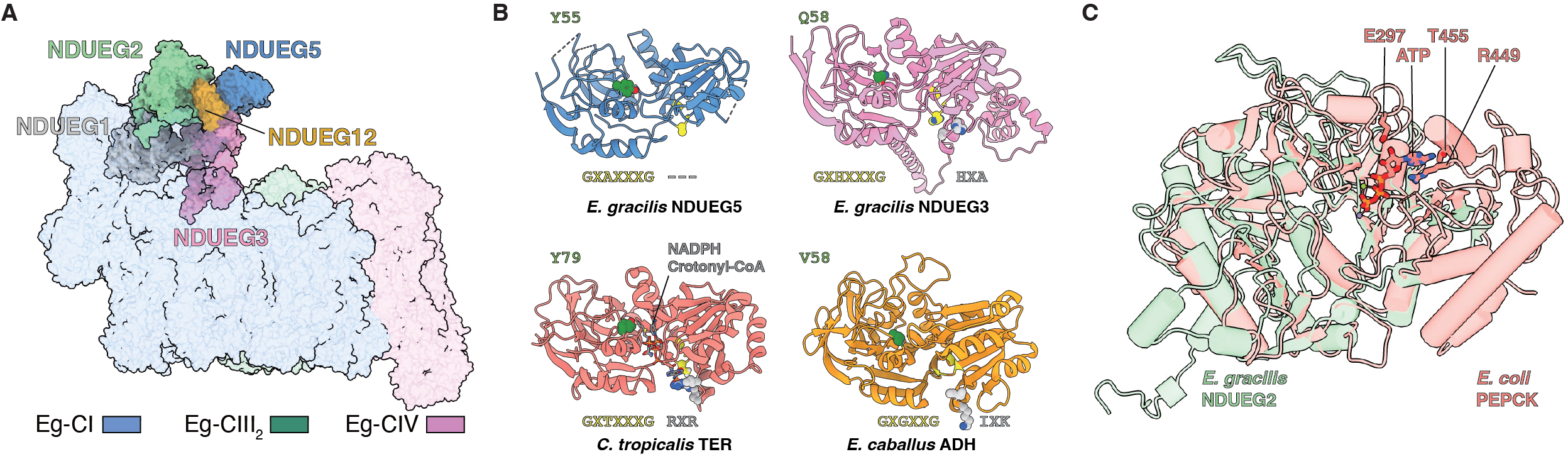
Another interesting point of E. gracilis’s ETC is that a ‘fatty acid synthesis (FAS)’ domain is found located on the very tip of Eg-CI PA (Figure 2A). This FAS domain has already been identified by mass spectroscopy-based study of Eg-ETC and located to CI PA via negative stain electron microscopy6. Our study provides the first high resolution picture of such domain and identified its five component subunits. Sequence and structural BLASTs reveal that these subunits are homologous to enzymes involved in the reversal process of fatty acid b-oxidation, in particular the NADH-dependent trans-2-enoyl-coenzyme A (CoA)/acyl carrier protein (ACP) reductases that convert 2-enoyl-CoA/ACP to its saturated form (Figure 2B). Another subunit in the Eg-FAS domain is structurally homologous to phosphoenolpyruvate carboxykinase (PEPCK) (Figure 2C), which may also contribute to the fatty acid synthesis/wax fermentation process of E. gracilis by supplying its precursor, malonyl-CoA, via fumarate and the anaerobic ETC CI-rhodoquinone-CII. However, despite thorough activity assay efforts, none of these FAS subunits has the activity of their homologous FAS enzymes. Such phenomenon is not unexpected in the field of ETC, since the previously reported g-carbonic anhydrase domain of non-opisthokont CIs, the short-chain dehydrogenase (SDR) homologous subunit NDUFA9 and the nucleoside kinase subunit NDUFA10 of mammalian CI are all either not active or cannot proven to be active3,7.
Why does ETC complexes collect these extra subunits homologous to random enzymes that cannot add new function besides electron transport? One hypothesis is that such subunits use to be active when they were added to ETC under adaptive selection pressure but have lost their activities later during evolutionary history. Another more likely scenario is, these subunits are added to compensate for the streamlining process of the mitochondrial genome during its co-evolution with the nuclear genome8. Loss of certain structural elements during mitochondrial genome simplification or splitting of certain gene to ease the process of nuclear translocation are both asking for ‘patching’ of the original structures disturbed9. The best candidate to fulfill this role could be some random enzyme in the vicinity with relatively high abundance at the time this happened, such as the g-carbonic anhydrase or FAS enzymes. Although seems less exciting, such non-adaptive narrative of the complexification process of ETC, increasingly seen for non-opisthokonts mitochondria, is gaining its popularity.
References
- Vercellino, I. & Sazanov, L. A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol23, 141–161 (2022).
- Zhou, L., Maldonado, M., Padavannil, A., Guo, F. & Letts, J. A. Structures of Tetrahymena’s respiratory chain reveal the diversity of eukaryotic core metabolism. Science (1979)376, 831–839 (2022).
- Maldonado, M., Guo, F., Letts, J. A. & Carter, A. P. Atomic structures of respiratory complex III2, complex IV, and supercomplex III2-IV from vascular plants. Elife10, 1–34 (2021).
- Letts, J. A., Fiedorczuk, K. & Sazanov, L. A. The architecture of respiratory supercomplexes. Nature537, 644–648 (2016).
- Han, F. et al.Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae. Nat Commun14, 2542 (2023).
- Miranda-Astudillo, H. V. et al.The atypical subunit composition of respiratory complexes i and IV is associated with original extra structural domains in Euglena gracilis. Sci Rep8, 1–13 (2018).
- Padavannil, A., Ayala-Hernandez, M. G., Castellanos-Silva, E. A. & Letts, J. A. The Mysterious Multitude: Structural Perspective on the Accessory Subunits of Respiratory Complex I. Front Mol Biosci8, 1–33 (2022).
- Huynen, M. A., Duarte, I. & Szklarczyk, R. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochimica et Biophysica Acta - Bioenergeticsvol. 1827 Preprint at https://doi.org/10.1016/j.bbabio.2012.08.001 (2013).
- Petrov, A. S. et al.Structural patching fosters divergence of mitochondrial ribosomes. Mol Biol Evol36, (2019).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

.png)





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in