Artificial Intelligence for Structural Modification of Natural Products
Published in Cell & Molecular Biology and Computational Sciences

Natural products (NPs) with novel chemical structures and diverse bioactivities are a vital source for innovative drug discovery. However, it often consumes numerous sample resources using traditional methods to modify their structures and obtain lead molecules with ideal druggability. The high costs and low efficiency inherent in this process pose a key bottleneck to the conversion of NPs into natural drugs.
What are Molecular Generation Models?
In recent years, AI drug discovery & design (AIDD) has provided innovative solutions for NPs structural modification research. As an important branch of AIDD, the molecular generation models have great potential in NPs structural modification based on “group modification” and “scaffold hopping”. Depending on the availability of known biological targets for NPs, generative models fall into two distinct categories:
- Target-interaction-driven:focusing on the use of structural information of the known targets to guide targeted structural modification
- Molecular activity-data-driven: in the case of unknown targets, predicting the potentials and guiding structural modifications based on the structure and activity data of known active molecules.
Numerous molecular generative models have emerged in recent years. For functional group modification, representative frameworks include DeepFrag, FREED, and DEVELOP; for scaffold hopping, DeepHop, SyntaLinker, and ScaffoldGVAE are notable examples. Most of the models are open-source, providing valuable computational resources for accelerating structure-based drug discovery.
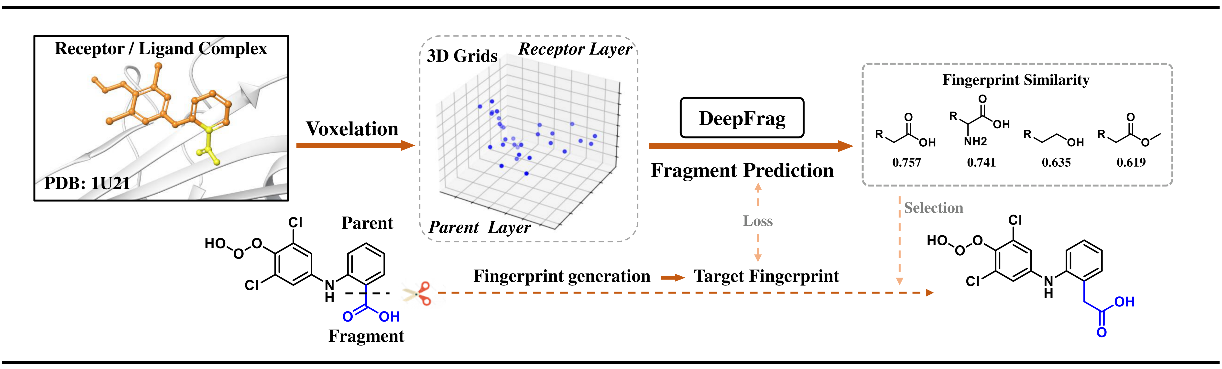
Figure 1 The workflow of DeepFrag
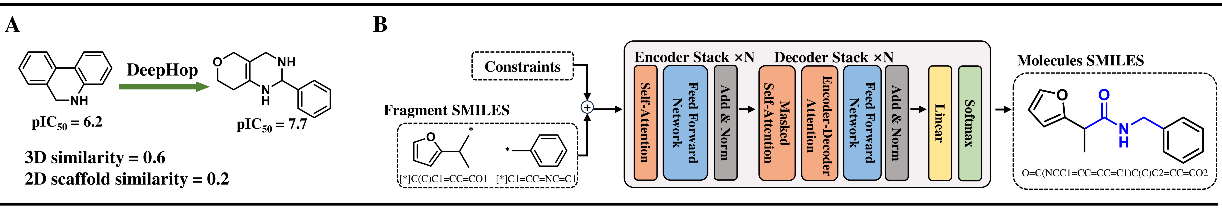
Figure 2 The workflow of DeepHop and SyntaLinker
How is Molecular Generation Models Applied in Structural Modification?
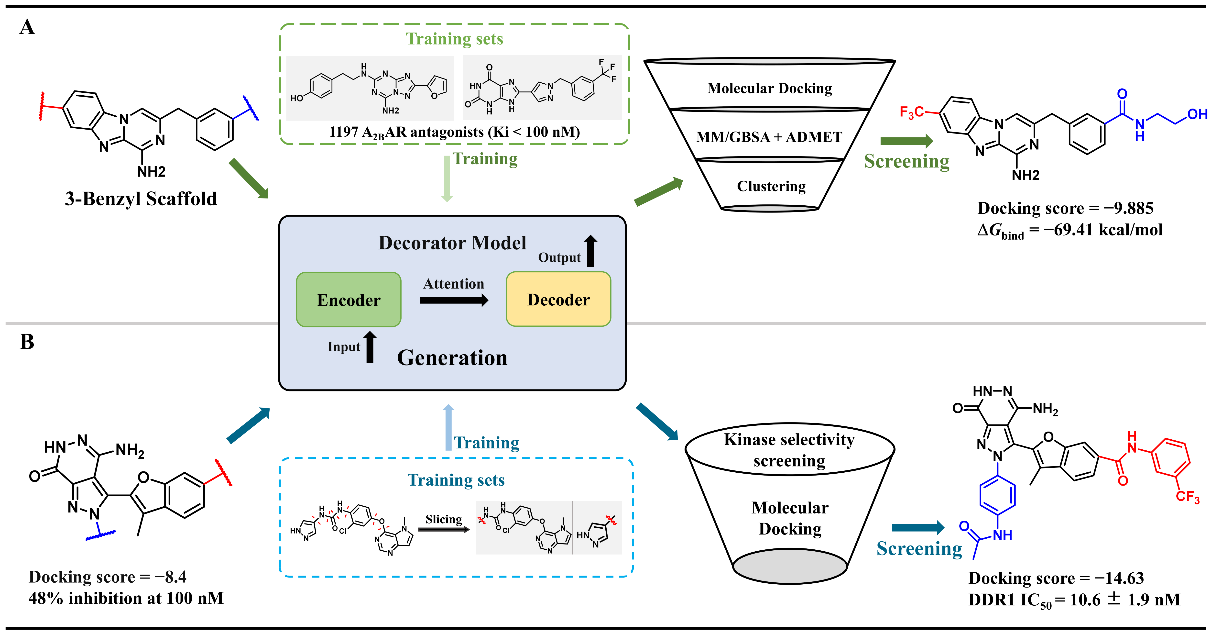
These generative models not only demonstrate significant technical innovation but also exhibit great value in practical applications, especially at the local and global levels of molecular optimization. For example, DeepFrag leverages protein-molecule interaction data to accelerate the development of anti-SARS-CoV-2 lead compounds (Figure 3A) and the optimization of Topo IIα inhibitors for enhanced anticancer potency (Figure 3B). Similarly, Scaffold Decorator integrates bioactivity data with various derivatization strategies to facilitate the discovery of highly selective adenosine A2B receptor antagonists (Figure 4A) and novel DDR1 inhibitors (Figure 4B).
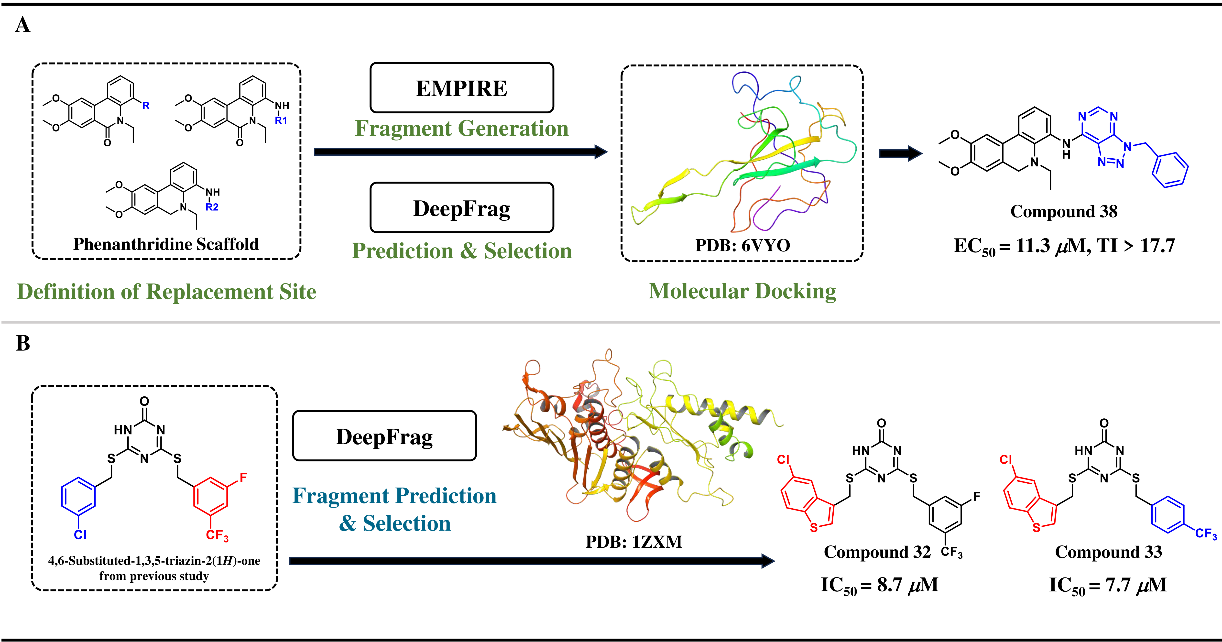
Figure 3 Two application examples of DeepFrag
Challenges and Future Directions
Despite significant advances in molecular generative models for natural product (NP) structural modification, critical challenges persist:
- Target-interaction-driven models rely on high-quality protein-ligand complex data(scarce and costly), with their predictive capabilities limited by dependence on prior target information, poor generalization to new or cross-species targets, and difficulty in simulating target dynamics (e.g., allostery effects, microenvironmental influences).
- Activity-data-driven models aresusceptible to dataset bias and experimental noise, lack mechanistic interpretability, and have insufficient chemical space coverage for complex NPs and new synthetic molecules.
- Core challenges include high computational costs, inadequatemodeling of multi-scale biological complexity, limited interpretability of generative outputs, unaddressed ethical risks, and persistent reliance on wet-lab validation for synthetic feasibility assessment.
Systematic breakthroughs in data, algorithms, and technologies, which remain imperative to overcome the challenges of data scarcity, multi-objective conflicts, and synthetic feasibility. Such advancements are critical for accelerating the bench-to-bedside translation of NPs from chemical entities into clinical drugs. In the future, technological innovations like lightweight model architectures, dynamic interaction modeling, and multi-modal data fusion are expected to overcome the key challenges of data insufficiency and low synthetic feasibility. By establishing an exclusive database of NPs and upgrading it to an intelligent prediction platform, and integrating deep learning design, automated synthesis and high-throughput screening, a closed-loop optimization system of “virtual design → robotic synthesis → experimental feedback” is being created, which will promote the optimization of NPs to a new level.
These findings were published in Natural Products and Bioprospecting in the review article titled "Bridging chemical space and biological efficacy: advances and challenges in applying generative models in structural modification of natural products" (https://link.springer.com/article/10.1007/s13659-025-00521-y).
Follow the Topic
-
Natural Products and Bioprospecting

This is a single blind peer-reviewed open access journal that devoted to rapidly disseminate research results in all areas of natural products.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in