Artificial Intelligence in Colonoscopy
Published in Bioengineering & Biotechnology
The paper in Nature Biomedical Engineering is here: go.nature.com/2CCOqXA
Colorectal cancer is the leading cause of cancer-related deaths worldwide. Colonoscopy could reduce the incidence and mortality of colorectal cancer via the detection and removal of precancerous polyps. Evidence has shown that each 1.0% increase in adenoma detection rate could result in a 3.0% decrease in the risk of interval colorectal cancer. Despite of the application of modern optic and digital technologies to colonoscopy, polyps can still be missed, with reported miss rates of up to 27%. Working in a high-volume endoscopy center in China, we could often find additional polyps that were missed in a recent colonoscopy procedure when doing polypectomy or endoscopic mucosa resection surgeries.
In 2015, I read a review paper ‘Computer vision and augmented reality in gastrointestinal endoscopy’ by Dr. Tyler Berzin’s team at Beth Israel Deaconess Medical Center. In this paper, the reasons for a missed polyp were thoroughly discussed: 1) It was never in the visual field; 2) It was in the visual field but not recognized. Innovations of optic hardware have sought to address the first problem by expanding the visual field and enhancing the definition, but it could not help with the second problem. Some evidence has shown that with trainees or nurses observing the video monitor, the polyp detection rate could be increased. Because of the inter- and intra-observer variability, these efforts remain controversial. Ideally, a real time automatic polyp detection system with both high sensitivity and specificity comparable to expert endoscopists, could serve as a more effective second observer.
I was so excited by the idea of real-time automatic polyp detection during colonoscopy that I discussed with my best friend Dr. Xiao Xiao, a mathematician who had extensive experience of modeling financial market using advanced mathematical models. After studying various colonoscopy cases in our center and basic morphological features of colon polyps, Dr. Xiao thought this goal was achievable. Then, I got fully support from directors of our GI division, Dr. Liangping Li and Dr. Xiaogang Liu.
As a first step, per Dr. Xiao’s request, I selected some colonoscopy images from the database of our center, and formed a panel of experienced endoscopists to label each polyp appearance. Then, Dr. Xiao used conventional deep learning method to build an algorithm. However, it was disappointing that the sensitivity and specificity were both around 70%, which was apparently insufficient for clinical application. After a thorough discussion on the results, we found that given significant intra-class variations and subtle inter-class variations for colon polyp and non-polyp structures, conventional machine learning and training methods, which tend to converge to a model that relies mainly on overall morphology, were ineffective. We found that an experienced endoscopist may rely ultimately on local structures, for example the pits pattern and micro-vessels, to confirm whether a suspected protruding object is a polyp. Then we focused on local features of polyps when preparing the training data and took specific consideration during the algorithm development. The resultant algorithm turned out to be quite satisfactory.
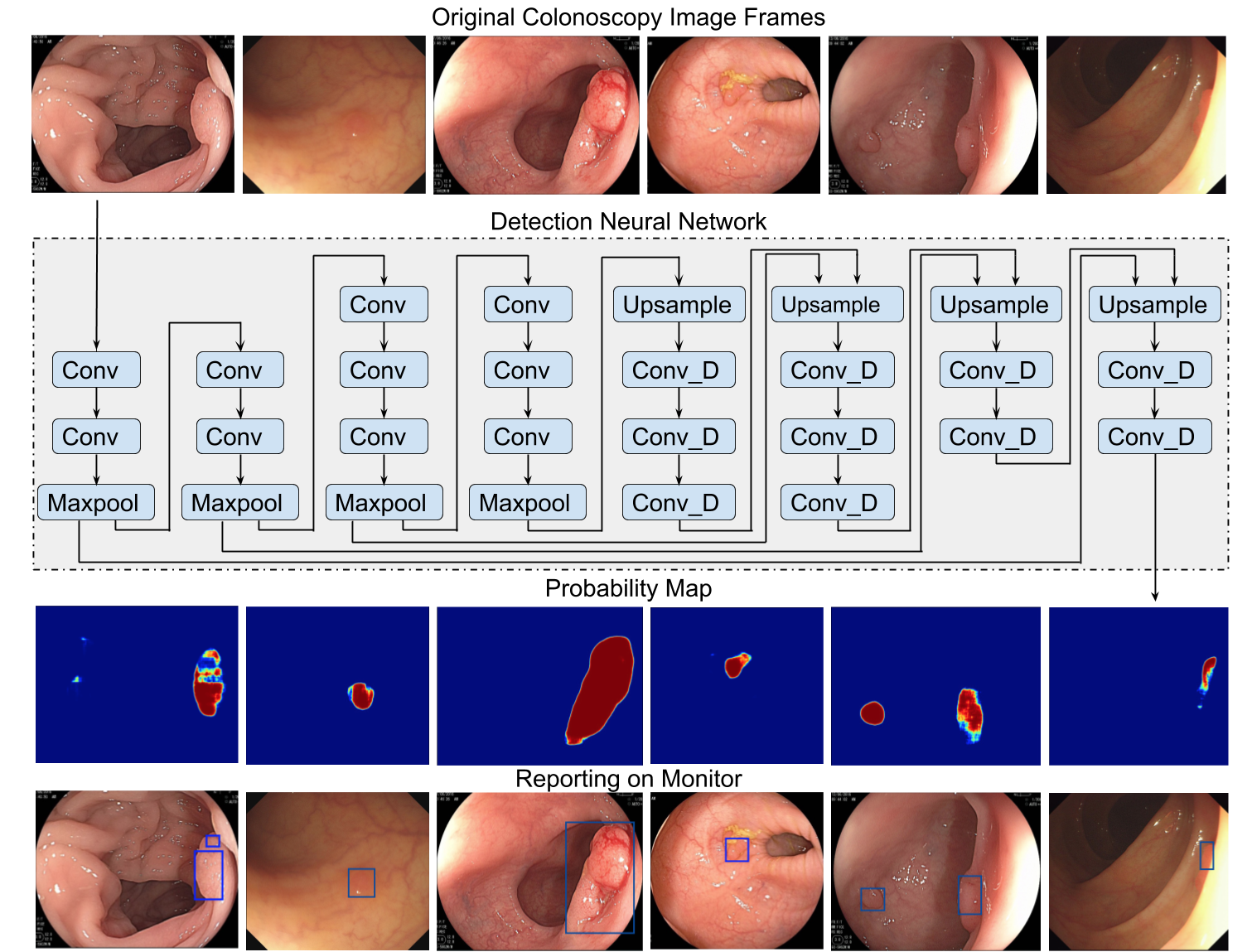
We validated the new algorithm on colonoscopy images collected prospectively with respect to training data. On this dataset of more than 1500 histologically different polyps, the algorithm achieved a sensitivity and a specificity both around 95%. We presented the abstract of this validation in the World Congress of Gastroenterology (WCOG) 2017 and was recognized as International Award.

As a next step, Dr. Xiao’s team integrated the algorithm into a real time computer aided diagnostic (CAD) system. When the system was first deployed in our center in Apr 2017, the latency was about a few hundred millisecond. With parallel deployment of Nvidia Titan X Pascal GPUs, the systems latency was shortened to dozens of millisecond at a 30 frames per second refresh rate. As a result, the endoscopist could perform colonoscopy only by looking at the monitor of CAD system, instead of the original monitor.
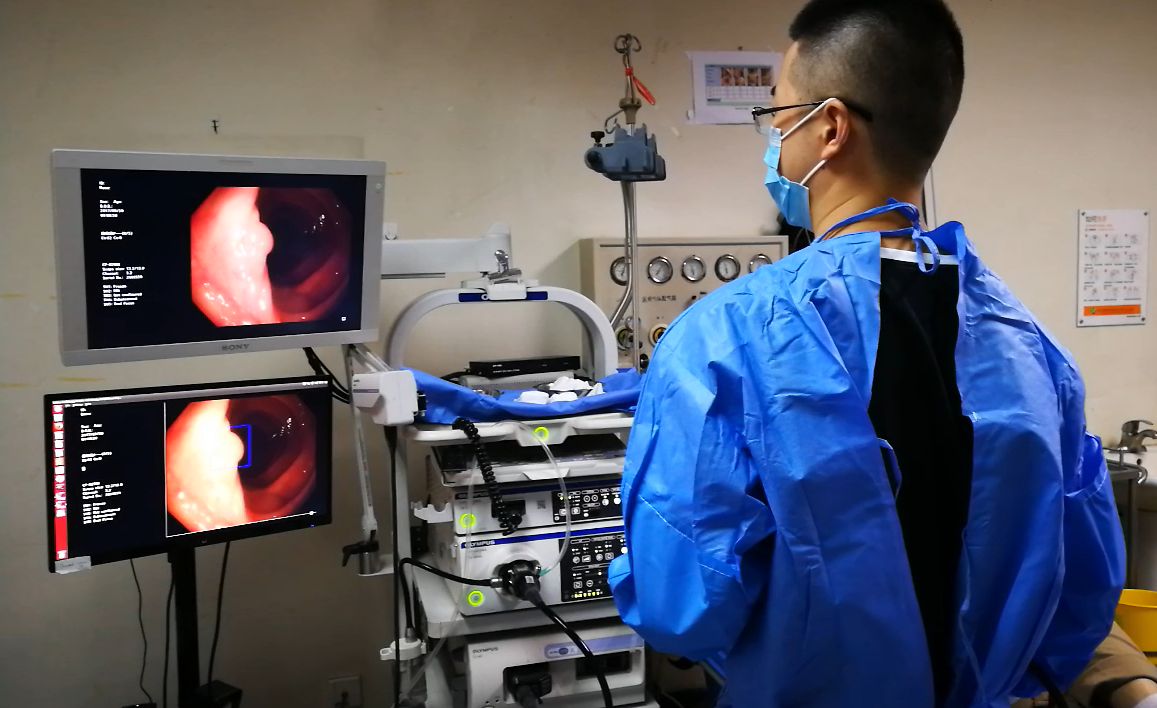
In order to avoid the bias caused by single-center sampling, we sent the system to Dr. Tyler Berzin at Beth Israel Deaconess Medical Center (BIDMC) of Harvard Medical School, and Dr. Berzin has observed a similar performance at his center. Then, we invited Dr. Berzin’s team to join our study on further validation of this CAD system.
To further investigate by how much colon polyp and adenoma detection rates can be increased with the assistance of this CAD system, compared to standard colonoscopy, we have accomplished a randomized controlled clinical trial. We have presented the interim results of this trial at Digestive Disease Week (DDW) 2018 and will present the complete results at United European Gastroenterology Week (UEGW) 2018.
In order to further validate the effectiveness of the system and hopefully to set it as a clinical routine in the near future, we also planned a double-blind clinical trial and a multi-center tandem study.
Given the overall high performance and real-time detection capability, this system may be useful in the real-life clinical setting in two ways. It could serve as a ‘second observer’ to report possible polyps that an endoscopist may have missed, so that to decrease the incidence of colorectal cancer. On the other hand, this system may serve as a quality-control metric when used for re-examination of colonoscopy videos after colonoscopy in order to assess the quality and level of doctors and trainees.
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in