Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria
Published in Bioengineering & Biotechnology

Motivation
The next time you’re driving your car over a bridge, take a moment to ponder how engineers designed both the car and the bridge on a computer, using predictive models to ensure that they work as expected across diverse real-world conditions (more than 99.99% of the time). Yes, automotive and civil engineers routinely build and test prototypes, but that data improved their overall discipline, refining their knowledge of the physical & chemical interactions controlling everything from air flows to material properties. As those disciplines matured, prototypes were no longer needed; when was the last time you saw someone build an at-scale prototype of a modern bridge?
If we endeavor to engineer biology without repeated trial-and-error experimentation – the oft-named design-build-test-learn cycle – Synthetic Biology needs to develop predictive design capabilities, encompassing all the physical & chemical interactions controlling biological functions. In our recent work entitled “Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria”, we take yet another step forward towards making that vision become a reality, with both conceptual and quantitative advancements that can be directly “plugged in” to our growing toolbox of system-wide design algorithms for engineering organisms.
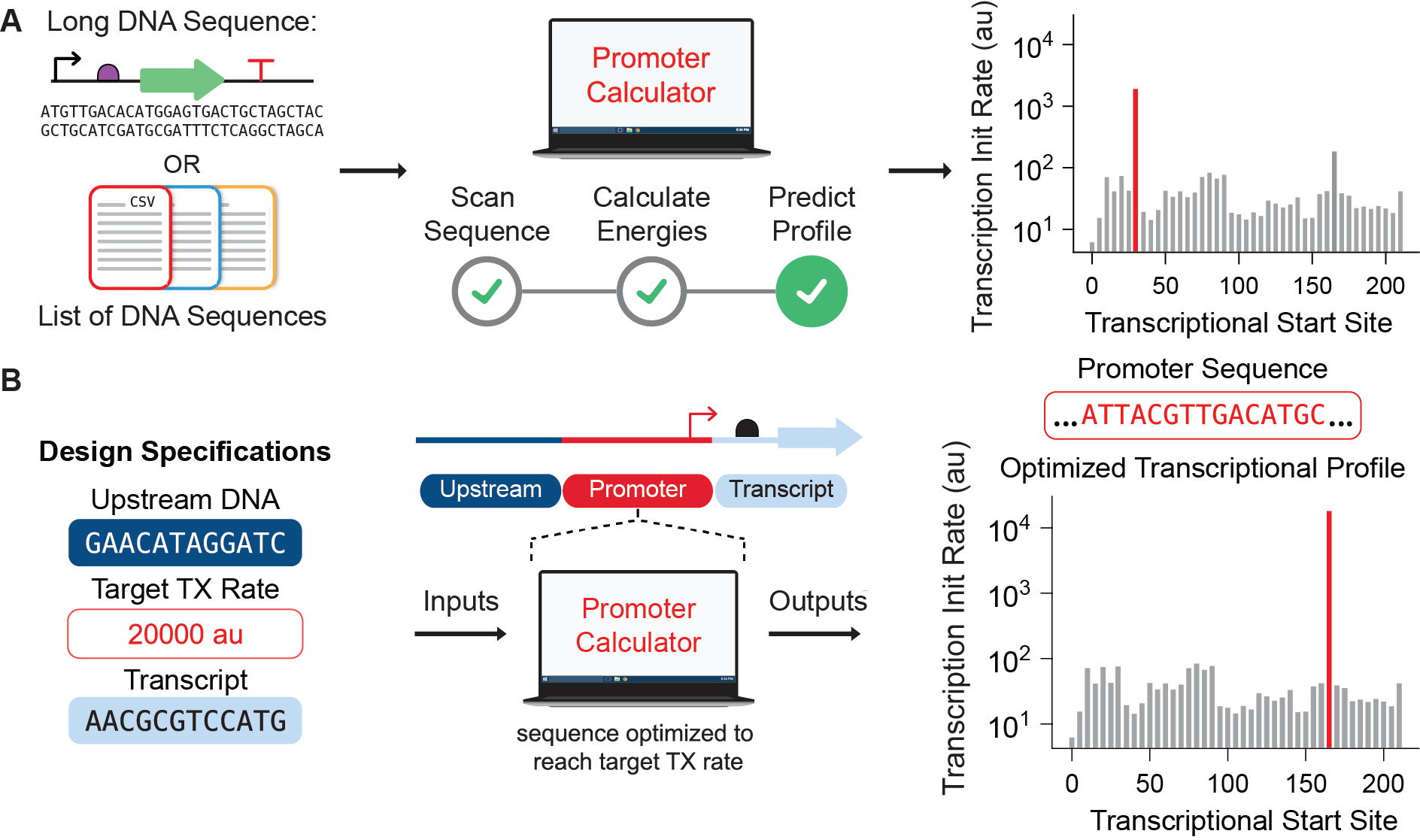
Figure 1: The Promoter Calculator in Predict and Design modes. (A) In Predict mode, the Promoter Calculator scans across a long DNA sequence and predicts the transcription initiation rate at every potential transcriptional start site, by calculating the thermodynamic free energies between RNAP/σ and each potential DNA binding site. (B) In Design mode, the Promoter Calculator designs a synthetic promoter sequence with a single predominant transcriptional start site and a targeted transcription rate, inputted on a proportional scale. The upstream DNA sequence and the mRNA transcript sequence are also inputted as they will also alter the promoter’s transcription rate, which is included in the model.
The Promoter Calculator
In a nutshell, here’s what we accomplished. We designed and characterized 14206 synthetic promoters in vitro by measuring their transcriptional start sites (TSSs) and transcription rates using bacterial RNAP/σ70 (the predominant transcriptional machinery during exponential growth). Using this data, we then trained and tested a biophysical model of transcriptional initiation (called the Promoter Calculator) by leveraging machine learning to determine the energetic contribution for each interaction between RNAP/σ70 and promoter DNA. We then validated the Promoter Calculator’s predictions across 16741 additional promoters that were characterized in a variety of in vivo conditions (different reporters, plasmid copy numbers, with vs. without self-cleaving ribozymes, measurement techniques, and more). We then applied the Promoter Calculator to design 35 synthetic promoter sequences with targeted single-TSS transcription rates across a >1000-fold scale. Finally, we utilized the Promoter Calculator to scan across a long genetic circuit sequence and predict the transcription rates of both the expected promoters controlling regulator expression as well as the unexpected promoters (e.g. inside coding sequences) that inadvertently disrupt genetic circuit function. Overall, we found that the Promoter Calculator accurately predicts site-specific transcription initiation rates in both “predict mode” and “design mode” (about R2 = 0.80).
Beyond this article, we added the Promoter Calculator to our user-friendly design platform for engineering organisms, available at https://salislab.net/software. The Promoter Calculator has already been used over 20,000 times by over 500 researchers to predict the transcription rates of existing genetic systems and to design synthetic promoters with desired transcription rates. In this blog post, we highlight three major conceptual and quantitative findings that could be useful in your own work.
Clean Data Makes a Good Model
With enough data, you can train a machine learning model to find a pattern, even if that pattern is not important, already well-known, or just plain meaningless. Our role as scientists & engineers is to acquire the best possible dataset to answer the novel questions that we care about, leveraging machine learning as a tool in that process. In this work, our objective was to develop a model to predict transcription initiation rates from any DNA sequence even when a promoter uses highly non-canonical motifs. We explicitly did not want to develop some Frankenstein model that empirically lumped together other gene expression processes (e.g. mRNA decay or translation). To do that, however, we needed to ensure that our experimental assays were measuring only changes in transcription rate without the possibility of other gene expression processes interfering and confounding those measurements. We overcame that challenge by carrying out in vitro transcription assays using only four ingredients: [1] our designed plasmid library (naturally supercoiled) with 14206 different promoters transcribing uniquely barcoded mRNA transcripts; [2] purified RNAP/σ70; [3] NTPs; and [4] aqueous buffer. This simple reaction contains no RNases, no ribosomes, and no other transcription factors or enzymes. When we applied RNA-Seq and DNA-Seq (with high read depth and in triplicate) to measure mRNA levels per promoter copy, we know explicitly that there is no opportunity for other gene expression processes to interfere with our transcription rate measurements. This is our “ground truth” on which to build a good model.
Transcriptional Profiles are Vectors and They Control Biological Function
The word “transcriptional profile” in our work’s title is important. In engineering disciplines, there are velocity profiles, temperature profiles, and concentration profiles. A profile quantifies how properties (velocity, temperature, or concentration) change along a particular length-scale. They are vectors and the numbers inside the vector are collectively governed by physical processes. For example, the temperature at two neighboring points affect each other through heat transfer. The same is true for transcription. Our Promoter Calculator predicts the transcriptional profile of a long DNA sequence; where and how much RNAP/σ70 binds to each site in that DNA sequence. If we want to understand holistically how a genetic system works, we can’t focus on the small DNA sequence regions that we call promoters. We need to consider the whole transcriptional profile of the system. For example:
[1] Strong natural promoters often have multiple transcriptional start sites (e.g. ribosomal RNA promoters), each with high transcription rates. Those distinct RNAP/σ70 binding sites may be regulated by different transcription factors, leading to complex multi-input gene regulation. The Promoter Calculator predicts the transcription rates from each distinct RNAP/σ70 binding site, enabling identification of multi-factorial gene regulation.
[2] Synthetic promoters with multiple transcriptional start sites will produce multiple mRNA transcript isoforms with distinctly different translation rates. When introducing mutations, it can be challenging to separately control multiple transcription and translation rates at the same time. It’s better to design synthetic promoters with only a single predominant transcriptional start site, using the Promoter Calculator in design mode, to provide more precise control over expression levels.
[3] As we show in our work, promoters can appear anywhere inside a genetic system, including within coding sequences. These promoters may transcribe in either direction, for example, leading to always-on expression of a regulator that must be tightly controlled (increasing leakiness) or repressing expression of a desired protein (anti-sense transcription). Using the Promoter Calculator, you can redesign your genetic system to avoid the accidental introduction of internal promoters, particularly inside protein coding sequences.
[4] There are system-wide interactions that control transcription rates, for example, changes in supercoiling density, anti-sense RNA interference, and transcription factor regulation. The net transcription rate of a mRNA transcript will be governed by the cumulative effects of these interactions. It is now possible to directly input the Promoter Calculator’s predictions into such system-wide models to predict these system-wide effects, which is a topic of future studies.
Using Machine Learning without Abusing It
While machine learning is a powerful approach to finding patterns in large amounts of data, there is a tendency to apply deep learning approaches (e.g. stacked convolutional neural networks or transformers) even though they have significant limitations. They require huge datasets (millions of datapoints) and deriving meaning from model coefficients remains an open challenge. In our work, our objective was to develop a generalized & predictive model with physically meaningful numbers, specifically, the thermodynamic free energies for each interaction between RNAP/σ70 and a DNA binding site. We desired a predictive model that made both “external predictions” (transcription rates) and “internal predictions” (binding energetics) that could be cross-compared to other independent measurements. We view these goals as extremely important towards developing a unified biophysical model of gene expression; each model must use the same physiochemical “language” without resorting to non-interpretable empiricism.
We achieved our goals by first formulating a biophysical model of transcriptional initiation (using statistical thermodynamics) to calculate how much Gibbs free energy is released when RNAP/σ70 binds to a promoter, creates a transcriptional bubble, and stabilizes it by forming an R-loop, which are the key steps before transcription initiates. We derived a multi-term free energy model that sums together the Gibbs free energy contributions from each individual interaction. Our biophysical model is simply a linear sum of free energies, one for each interaction that takes place. We then make the quantitative connection between our free energy model and a classic type of machine learning (linear regression models with regularization) that use a linear sum of variables to predict an outcome. Here, the variables are either categorical (a binary variable, indicating the presence/absence of an interaction between RNAP/σ70 and a DNA binding site) or numerical (a calculated biophysical characteristic, e.g. the thermodynamic stability of the R-loop). Each of these variables is associated with a coefficient with an initially unknown value. The objective of the machine learning is to identify the optimal values of these coefficients so that, when the coefficients are multiplied by their respective variables and summed together, the predicted outcome is closest to the actual outcome (the change in Gibbs free energy). The optimization process also includes regularization, which adds different types of penalties to prevent over-fitting or misbalancing of coefficient values.
When coupled to a rigorous training & validation workflow (10-fold cross-validation) with final testing on unseen data, this approach worked spectacularly. Our Promoter Calculator was highly accurate on our collected dataset and generalized well across 16741 additional promoters from other datasets. But, most importantly, the model’s learned coefficients are physically meaningful and human understandable. The model training correctly quantified the strengths of the canonical interactions controlling σ70-dependent transcription initiation (e.g. TATAAT and TTGACA hexamers), while also quantifying the strengths of all other non-canonical interactions on the same energetic scale. With these energetic coefficients in hand, we don’t even need the machine learning model framework to make transcription rate predictions. We scan across long DNA sequences, enumerate potential binding sites, use the coefficients to calculate the interactions’ Gibbs free energies, sum them together, and use Boltzmann’s classic relationship to relate free energies to binding probabilities. In fact, we don’t even need a computer to design synthetic promoters; our motif energy look-up table and a pocket calculator would suffice. The deep learning revolution is cool, but let’s not overlook our human brain’s ability to deeply learn and come up with simpler, more interpretable, and more physically meaningful models.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in