
Six years ago, when our group first started discussing the need for continuous phylogenetic analysis of whole-genome sequenced bacterial isolates, large-scale meant hundreds to thousands of samples and not hundreds of thousands to millions, as it does now. Whole-genome sequencing (WGS) was gradually making its way into the toolbox of public health laboratories, having been proven useful to detect and track outbreaks of food-borne bacterial illnesses and other infectious diseases. WGS is now part of the routine in many public health, clinical and food safety microbiology laboratories, and thousands of sequences are being shared publicly every week. Hence, our “outbreak-spotting” pipeline, called evergreen, has gone through a long development phase, to tackle the new challenges.
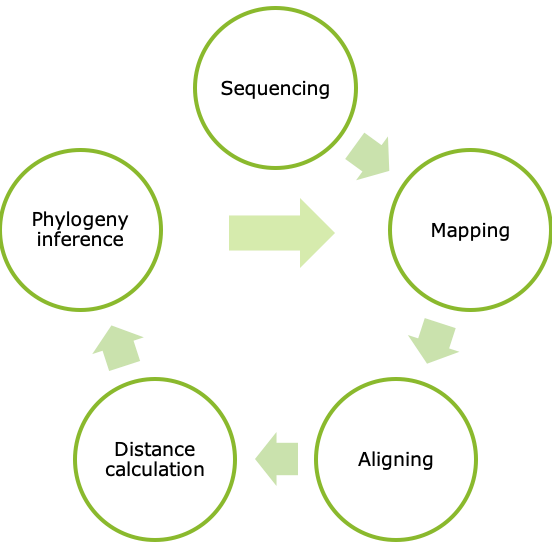
In a regular phylogenetic analysis, all sequences in the study are compared with all other sequences in the study. The computational cost of this is increasing with the square of the number of sequences in the study. One could see, that it would become unfeasible to compute when the number of sequences became too large. Furthermore, a phylogenetic tree is made on the sequences available at given time, and when new sequences arrive, it is often a wish to add these to the already available ones. The challenge was to do this in a fast and accurate way.
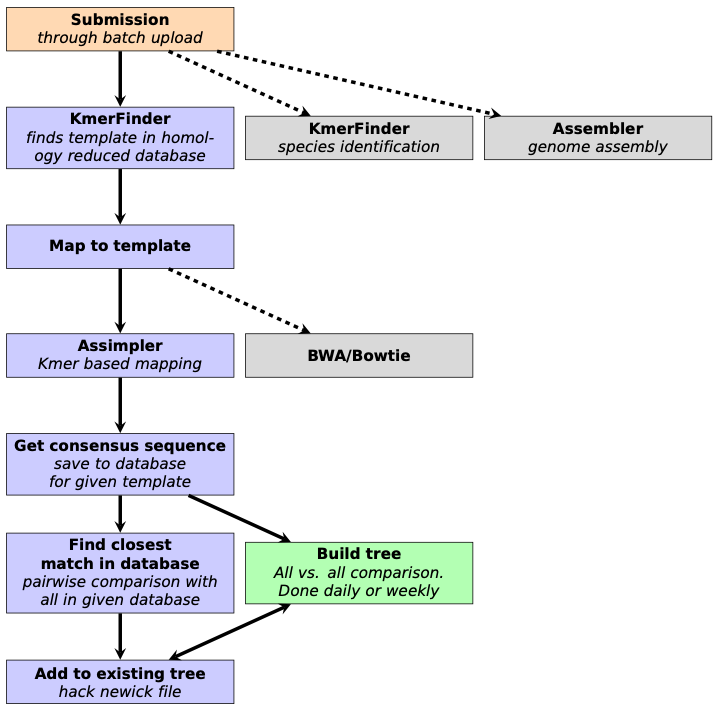
From the get go, the key idea for reducing computational burden was to divide the problem into many, smaller problems, or in this case, phylogenetic trees. In reference-based phylogenetic inference, the closer the reference is to the studied sequences, the better results can be achieved. Thus, splitting up the sequences by their sequence identity to selected reference sequences was an evident move. These references were chosen by homology reducing complete chromosomal genomes, to lessen the overlap between trees. The threshold is by default set to 1% nucleotide sequence difference on the whole genome. In the beginning, k-mer based identity calculations had been performed with the algorithm in KmerFinder1, which was later swapped out with KMA2, to speed-up the process by a hundred-fold.
The hope was, that these smaller trees would flourish in time, and in anticipation, a new genetic distance calculation method was implemented, that doesn’t necessitate the re-calculation of the full distance matrix each time a new sequence is added. This reduced the growth of the computational time of updating an existing “evergreen” tree.
Initially, the plan was to couple this pipeline to the Bacterial Analysis Pipeline running on the Center for Genomic Epidemiology website3, so users could monitor their own isolates for outbreaks. However, as more and more laboratories were publishing their WGS data in public repositories, the idea of the Evergreen Online platform emerged, which could connect food related samples to clinical samples, possibly revealing the culprit behind foodborne-disease outbreaks. Even across country borders! But this meant a lot more samples, than we originally planned for, so we added a homology-reduction step for the WGS samples. The threshold in Evergreen Online is 10 bases, which loosely corresponds to cut-offs for outbreak clusters, so we can use these groupings for surveillance as well.
These simple steps made it possible to compare hundreds of thousands of isolates since Evergreen Online started running, and we are planning further development to meet new demands.
References
1. Larsen, M. V. et al. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 52, 1529–1539 (2014).
2. Clausen, P. T. L. C., Aarestrup, F. M. & Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19, 307 (2018).
3. https://cge.cbs.dtu.dk/services/
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in