Behind the scenes: Microbiome-metabolite linkages drive greenhouse gas dynamics over a permafrost thaw gradient
Published in Microbiology, Protocols & Methods, and Sustainability
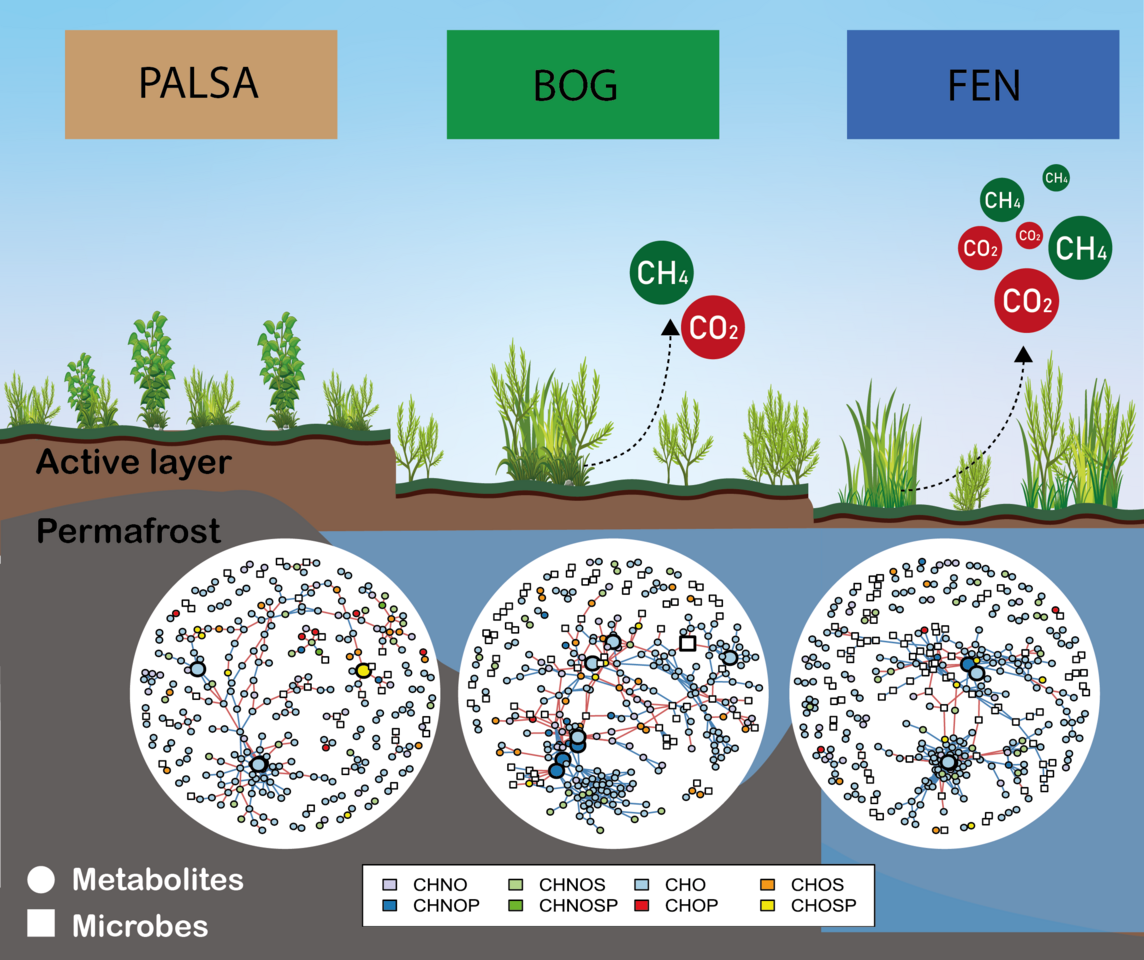
Thawing permafrost isn’t just about melting ice; it triggers a fundamental restructuring of the peatland environment.
Vegetation patterns shift, microbial communities adapt, and the very composition of the soil changes. Each of these elements play a crucial role in the production of greenhouse gases like CO2 and CH4. The critical question isn’t just how much carbon will be released, but in what form—will it be the less potent CO2, or the more powerful CH4? The answer lies in understanding the intricate interplay between the peatland’s living and non-living components.
In our study we focused on two key players in this unfolding drama: the microbial communities that inhabit these peatlands, and the peat metabolites —small chemical compounds—that serve as a chemical language, providing insights into microbial activities and environmental shifts.
Our Research Journey
Our study took us to Stordalen Mire, a thawing peatland located in Northern Sweden (68.35° N, 19.05° E). This site is a natural laboratory, showcasing different stages of thawing: intact (Palsa), partially thawed (Bog), and fully thawed (Fen) peat (Figure 1). What made this research truly special was the collaborative spirit behind it. As part of the National Science Foundation-funded EMergent Ecosystem Responses to ChanGE (EMERGE) Biology Integration Institute (Figure 2), we brought together a diverse team of experts, each contributing unique skills to our study of permafrost thaw: Field Team: Every summer, these dedicated researchers travel to Stordalen Mire to collect samples from the palsa, bog, and fen sites. Their hard work provides the foundation for our analyses. Microbial Ecologists: These scientists identify and understand which microbes are present in our samples, helping us understand their roles in peatland decomposition. Their work is crucial in linking microbial activity to ecosystem processes.; Soil Biochemists: with their expertise in physicochemical properties of peat, these team members help us uncover the factors controlling microbial decomposition. Their insights are vital in interpreting how soil chemistry influences microbial activity.; Data Scientists: Organizing and analyzing our extensive dataset, these experts use advanced techniques to integrate diverse data types. Their work is essential in revealing the complex interactions between microbial decomposition and climate feedback.

The Science Behind the Scenes
As part of this team, we at the Tfaily lab contributed to the study of the peatland metabolome by using advanced mass spectrometry (MS) techniques to analyze the chemical makeup of peat in each of the habitats along the thaw gradient. Using advanced mass spectrometry techniques, particularly Fourier-transform ion cyclotron resonance (FT-ICR) MS via direct injection, we created a chemical fingerprint of each sample. It is like CSI for ecosystems! For example, based on the number of carbon, hydrogen, and oxygen atoms on the molecular formula, we can assign metabolites to major biochemical classes (e.g., lipid-, protein-, lignin-, carbohydrate-, and condensed aromatic-like) or derive biochemical indexes that can provide further information about them, such as the nominal oxidation state of carbon (NOSC) that can tell us if they are more or less available for the microbes to use.
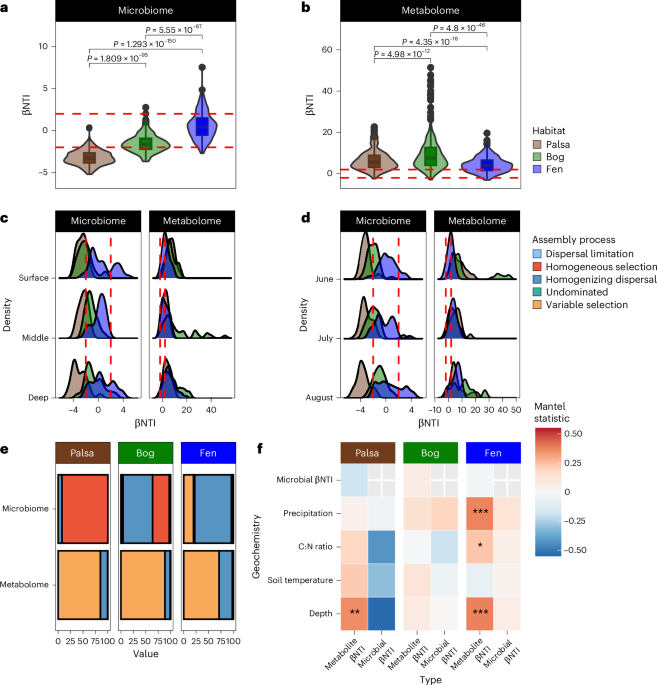
One of the most exciting aspects of our research was applying community assembly theory to both metabolites and microbes. This approach, typically used to understand how microbial communities form, allowed us to explore the factors influencing both living organisms and their chemical environment. It is a bit like trying to understand a conversation by listening to both speakers simultaneously. Specifically, in this framework, communities are formed through a mix of predictable (deterministic) and random (stochastic) processes. For example, communities can be shaped by environmental factors or interactions between species (selection), random changes in species abundance (drift), the emergence of new species (speciation), and the movement of species between areas (dispersal). By using null modeling approaches developed by Dr. James Stegen team, we can estimate how much each of these processes influence microbial community assembly. Furthermore, recent studies have found that this approach can also be applied to understand the factors influencing the formation of chemical profiles (metabolomes).

Figure 2: EMERGE Group Picture at Biosphere 2- 2023. Photo Credit: Kim Land.
What We Discovered
Our findings were intriguing. From a broad perspective we saw contrasting ecological processes shaping microbial and metabolite community profiles across the permafrost thawing gradient. But when we zoomed-in we found key microbial species and metabolite classes (particularly lignin-like metabolites rich in sulfur and nitrogen) that could be driving greenhouse gas emissions, especially in bog sites (Figure 3).These sulfur-rich compounds, likely derived from the abundant Sphagnum mosses in bogs, highlight the complex interplay between vegetation, soil chemistry, and microbial activity. It's a reminder that in ecosystems, everything is connected.
Why It Matters
As metabolites in a system reflect the past and the future processes, understanding them in tandem with microbial community shifts can help us find key patterns or blueprints that we can use to study and predict how ecosystems function and how they respond to disturbances, whether natural or anthropogenic. In a time of rapid environmental change, the discovery of complex connections that underpins ecosystems is more crucial than ever.
Looking Ahead
As we continue this research, I'm both excited and sobered by its implications. The potential for increased greenhouse gas emissions from thawing permafrost is concerning, but our growing understanding of these systems offers hope for developing strategies to mitigate these effects.
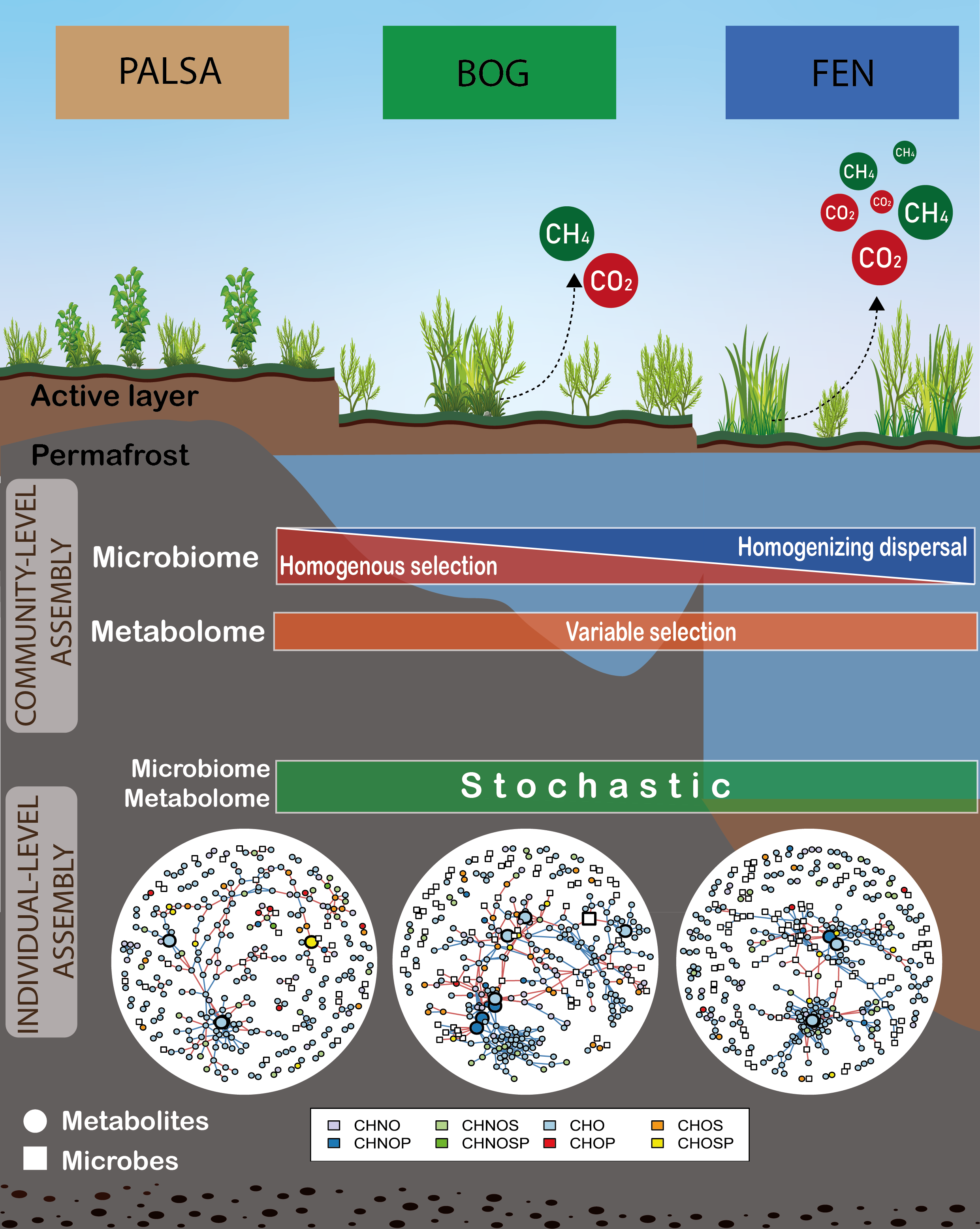
Figure 3: Visual abstract showcasing main results of the paper. Created by Viviana Freire-Zapata and Salome Freire-Zapata.
References.
- Tarnocai, C., J. G. Canadell, E. A. G. Schuur, P. Kuhry, G. Mazhitova, and S. Zimov (2009), Soil organic carbon pools in the northern circumpolar permafrost region, Global Biogeochem. Cycles, 23, GB2023, doi:10.1029/2008GB003327.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in