Beyond ATGC: enzymatic synthesis and nanopore sequencing of 12-letter supernumerary DNA to unlock xenobiology's potential
Published in Bioengineering & Biotechnology

Challenging the rules of Nature with unnatural base pairing xenonucleic acids (XNAs)
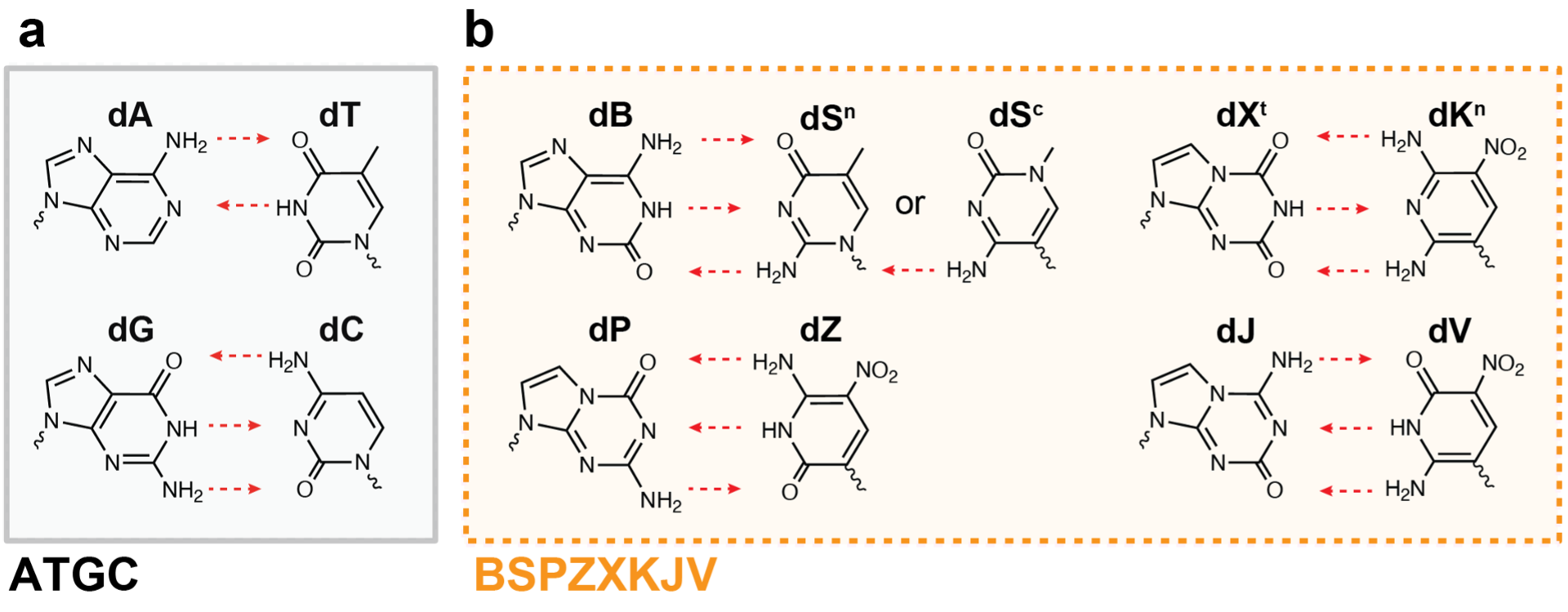
The 4-letter DNA code Nature utilizes (A, T, G, C) is the backbone of the central dogma and the very blueprint of life itself. Our ability to manipulate this code is also the driver of biotechnological progress; everything from genetically engineered organisms to therapeutics, diagnostics, and information storage relies on this 4-letter code. Yet, as biotechnology advances, science is beginning to transcend these rules set by Nature. Unnatural base pairing Xenonucleic acids (ubp XNAs) are synthetic, nucleotide analogs that can be used orthogonally to the 4 letters in DNA. While there are many types of ubp XNAs, one such variation involves up to 12 different nucleotides that can form 6 complementary hydrogen-bonding pairs. We term this nucleic alphabet soup ‘supernumerary DNA’ (Figure 1).
Ubp XNAs have the potential to revolutionize every aspect of biotechnology, from semi-synthetic organisms that have unimaginably large codons tables (1,728 codons with 12 letters rather than 64 codons with ATGC-only); to aptamer/aptazyme therapeutics that have novel binding modalities or reactivities; to ultrasensitive diagnostics; and even be the basis for storing digital information (i.e., files, movies, books) in DNA.
When we first opened our lab in January 2021, we were captivated by the possibilities of a world where working with arbitrary DNA alphabets were routine. However, we quickly realized that the abundant and crucial toolkits for 4-letter DNA technologies were not available for expanded DNA alphabets. As a new research group with limited resources, we hit our first road block in writing with these new letters: commercial options for XNA synthesis were limited and expensive (a few hundred dollars per base). The second hurdle we encountered came from reading. Commercial sequencing services and instruments could not sequence the ubp XNA letters we wanted to work with (B, S, P, Z, X, K, J, V). At this point, we connected with like-minded collaborators who also encountered similar problems. If we wanted realize this world with XNA aptamer therapeutics and XNA semi-synthetic organisms with an ATGCBS genome, we first had to lower these barriers to enter the field. We decided to focus our group’s first project on reading and writing with expanded letters.
Enzymatic synthesis of XNAs
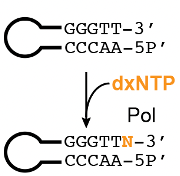
Our lab initially set out to find a synthesis solution that was applicable across all ubp XNAs we were working with, and accessible in both reagents and expertise. While chemical synthesis (e.g., using phosphoramidites) seems like a general solution, limited chemical stability had made routine organic synthesis challenging. Keeping these factors in mind, we found ourselves turning to an enzymatic synthesis solution using an old discovery, made over 35 years ago. Back then, scientists revealed that a fragment of E. coli DNA Polymerase I (Klenow Fragment exo-) could catalyze the addition of a single 2′-deoxynucleoside triphosphate (dNTP) to the free 3′-OH end of blunt-end double stranded DNA. This reaction, colloquially known as tailing, has an advantage over other enzymatic synthesis approaches as it favors a single base addition and does not require modified nucleotides or polymerase-nucleotide conjugates. We suspected that if a polymerase can tail all four standard dNTPs, it should be able to tail 2′-deoxy-xenonucleoside triphosphates (dxNTPs) as well (Figure 2). Our experiments confirmed this notion: a combination of analytical techniques including gel electrophoresis and high resolution liquid chromatography-mass spectrometry (LCMS) assays, we found that two polymerases (KF exo- and Therminator) were capable of tailing a single dxNTP onto blunt, dsDNA. Since tailing only incorporates one expanded letter, we needed to couple this reaction with a ligation step to make a true unnatural base pair. To incorporate a base pair, we next screened commercially available DNA ligases to ligate two hairpins with complementary ubp XNA overhangs. The hairpin construct becomes crucial here: unligated hairpins can be digested by a subsequent exonuclease step, which cannot target successfully ligated products as they lack a free 5′- and 3′-end. With these two steps, XNA tailing and XNA ligation, we now had a method for single XNA base pair insertion into DNA.
Nanopore sequencing of supernumerary DNA
Having successfully developed a method for single XNA insertion in DNA, we turned our attention to the other towering barrier: how to read these letters (or sequencing). Here, we focused on adapting existing technology rather than inventing a new one. We chose to use nanopore sequencing since this method is theoretically does not require special fluorophore-labeled bases to sequence. In nanopore sequencing, a voltage is applied across the membrane leading negatively charged DNA to traverse a pore, generating a small but measurable current signal. This current output as a function of the chemical structures of the nucleotides going through the pore. Oxford Nanopore Technologies (ONT) has made this platform accessible with their MinION sequencer, and with no need for modified nucleotides or additional equipment other than a computer, the missing link was model that could assign current signals to the correct XNA-containing sequences.
To fill this missing piece, we built XNA “kmer models”. In the kmer model of basecalling, the DNA current is only a function of the nucleotide going through the pore and its surrounding nucleotide context. Using XNA tailing and ligation, we built libraries that produced every 4-nt kmer possible containing an XNA; sequencing these libraries allowed us to build models that assign current signals to sequences, and subsequently basecall new sequences. To make all of our models accessible we also built Xenomorph, a reference-based basecaller available on Github that contains all of our measured models, which can perform end-to-end processing from raw nanopore data to basecalled results. We used Xenomorph to benchmark a validation set separate from our model building sequences and found that recall ranged between 60-87% when comparing each XNA to its most similar standard base; a consensus basecall of at least 10 reads increased this recall to 63-99%. What made this model building strategy most attractive was their low data requirement, meaning we could build models quickly and efficiently. With increased investment in data collection, including increasing complexity of libraries or increasing kmer size, we see a reasonable avenue for improving sequencing performance.
As an interesting and fun experiment to wrap up this project – we pushed our findings to their alphabetical limit. The inherent compatibility of XNA tailing and XNA ligation strategy with other DNA assembly strategies meant we could write, for the first time, 12-letter DNA. More so, the modularity of our sequencing models meant that we could apply them to also read 12-letter DNA. To do this, multiple ubp XNA-containing constructs underwent Golden Gate ligation to assemble two constructs that contained 4 standard letters (ATGC) as well as the additional 8 ubp XNAs (BSPZXKJV). We had built two version of this 12-letter supernumerary DNA sequences since we had two version of the S nucleotides (C-nucleoside and N-nucleosides). We named these sequences Scuper-12 and Snuper-12. Even in this complicated sequence space where each XNA was being compared against 11 other letters, we were able to properly decode all XNAs in Scuper-12. In Snuper-12, only Kn was incorrectly decoded, but this decoding error is easily resolved with additional priors.
The future of ubp XNAs, xenobiotechnology, and xenobiology
Altogether, this work establishes synthesis and sequencing methods that significantly lower the barrier to access XNAs in synthetic biology and beyond. In the advanced world of ATGC-based technologies, we understand the limitations of our work, including the fact that our basecaller is for sequence contexts where an XNA is embedded within a standard DNA context. However, we are hopeful that by laying down the first steppingstone, this work will encourage us and other groups to catalyze future XNA synthesis and sequencing innovations. In the immediate future, the methods we’ve developed can be used to study XNA retention in vivo, work with an expanded genetic code, develop aptamers with novel functions, and more. While XNAs are currently not as widely adapted as DNA, we’ve taken one step closer to a xenobiology world.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in