Interspecies interactions within microbial communities are complex, where both competition and cooperation can happen. Accurately mapping the population dynamics is critical for investigating function of complex microbial communities. The most conventional method is 16S rRNA gene amplicon sequencing analysis, which is prone to PCR bias. In our recent study, we explored a new approach to measure the copy numbers of 16S rRNA genes from different bacterial species directly by using the NanoString nCounter® system. This system is widely used to profile gene expression of eukaryotic cells with digital molecular barcodes. We have modified the digital barcodes to target the specific regions of 16S rRNA genes of 18 bacterial species from a mixed-species community. Our experiment showed that the NanoString nCounter® system can accurately examine the bacterial population dynamics without PCR amplification from microbial communities. Pseudomonas aeruginosa was found to be the most dominant species in biofilm community while not in planktonic community. Figure 1 shows the different cross sections of mixed-species biofilm community.
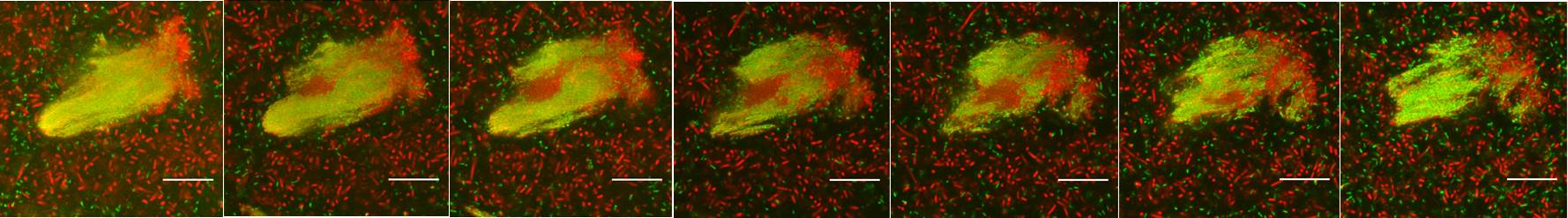
Figure 1. Confocal images of multiple species biofilm microcolony. Each section is around 2.8 µm steps perpendicular to the microscope optical axis. P. aeruginosa cells with GFP tag appears green; all of other bacterial cells appear red which were stained by Syto62; scale bars show 20 µm.
It is well known that P. aeruginosa is widely distributed in natural environment. It is also one of the leading pathogens for patients who suffer from cystic fibrosis and chronic wound infections. These infections are rather complicated which are characterized by co-existence of multiple bacterial species. However, most of the current understanding about P. aeruginosa physiology is derived from its in vitro and in vivo mono-species cultures, which might not reflect its real physiological condition in complex environment. The mixed-species community which consists of seventeen other bacterial species was thus used as a complex laboratory community model system to study the physiology and interaction of P. aeruginosa in multiple-species community.
To understand the mechanisms employed by P. aeruginosa to become the dominant species under biofilm cultivation condition, we compared its transcriptome profiles from mixed-species biofilms and mono-species biofilms using RNA-sequencing analysis. Our results showed that P. aeruginosa is able to gain fitness over other bacterial species under biofilm condition with the joint efforts of H1 type VI secretion system (T6SS), Psl exopolysaccharide and type IV pili. Psl exopolysaccharide and type IV pili are determining the P. aeruginosa biofilm formation in mixed-species biofilms while T6SS is a strong weapon for P. aeruginosa to target other species. Both confocal images and fluorescence based detection approached proved that the proportion of P. aeruginosa H1-T6SS null mutant in biofilm community decreased significantly compared to its parent PAO1 strain (paper figures). Our results suggest that biofilm formation might have a profound impact on physiology and function of complex microbial community such as microbiome.
Our paper can be found on the npj Biofilms and Microbiomes website: https://www.nature.com/articles/s41522-018-0076-z
Follow the Topic
-
npj Biofilms and Microbiomes

The aim of this journal is to serve as a comprehensive platform to promote biofilms and microbiomes research across a wide spectrum of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Natural bioactives, Gut microbiome, and human metabolism
Publishing Model: Open Access
Deadline: Feb 20, 2026
Harnessing plant microbiomes to improve performance and mechanistic understanding
Publishing Model: Open Access
Deadline: Jun 01, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in