Birth of a pathogen: The emergence of Kingella kingae virulence from the acquisition, co-option, and duplication of toxin-associated genes.
Published in Microbiology
The species in the Kingella genus are often found in the human oral and oropharyngeal microbiome. Some species in this genus, such as K. oralis and K. denitrificans, rarely cause invasive disease in individuals with intact immune systems. In contrast, K. kingae and K. negevensis are important causes of severe, invasive disease in otherwise healthy individuals and particularly young children. As diagnostic technologies improve, K. kingae is increasingly recognized as a major etiological agent of pediatric bone and joint infections.
Several virulence factors facilitate invasive disease by K. kingae and K. negevensis, including type IV pili, a capsular polysaccharide, a galactan-modified lipopolysaccharide, and an RTX-family toxin. Of the Kingella species, only K. kingae and K. negevensis (the pathogenic Kingella species) produce galactan and secrete the RtxA toxin (Fig. 1). RtxA is essential for cytotoxicity in vitro and for systemic disease in vivo. In this study, we sought to better understand the evolutionary events that led to toxin production and secretion by the pathogenic Kingella species.
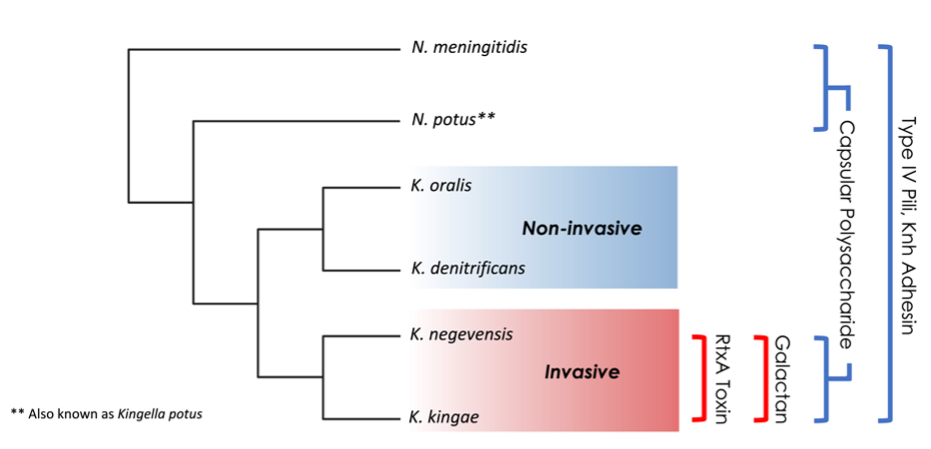
The RtxA toxin produced by K. kingae is post-translationally modified by RtxC, which is encoded by the rtxC gene immediately upstream of rtxA. The modified toxin is then shuttled out of the bacterial cytoplasm by a dedicated type I section system (TISS), comprised of RtxB, RtxD, and the outer membrane protein TolC. In many other bacteria, the genes encoding RTX-related proteins are found in a single operon, with the exception of tolC. By contrast, the rtx genes in K. kingae were originally reported to be present at two distinct locations in the genome. Using a large collection K. kingae isolates, we found that most isolates contain the genes encoding the toxin secretion system plus a single copy of rtxC at one locus and a second copy of rtxC, rtxA, and tolC at a second locus, such that the secretion system is separated in the genome from its toxic cargo. Genotyping showed that 30-35% of K. kingae clinical isolates have a duplicated copy of rtxC, rtxA, and tolC that is adjacent to the secretion system locus, resulting in two toxin-producing loci. Remarkably, we found that having two toxin loci was strongly correlated with invasive disease.
As K. kingae and K. negevensis are very closely related species and no other Kingella species have genes for the RtxA toxin, we hypothesized that acquisition of the rtx genes was a critical step in the evolution of these pathogens. Multiple lines of evidence pointed to a horizontal introduction for the toxin genes, including G+C% content, which is reduced for rtxCrtxAtolC relative to the rest of the genome, as well as gene-by-gene phylogenetic analysis. However, the secretion system genes rtxB and rtxD have a G+C% content that is very similar to the rest of the genome and have close homologs in non-pathogenic Kingella species. These data suggest that rtxCrtxAtolC was acquired independently. This system then co-opted pre-existing TISS components for toxin secretion.
To better trace the genomic rearrangements that led to the observed loci in the pathogenic Kingella species, we used whole genome sequencing and phylogenomics to show that the two loci likely fused in a common ancestor and are maintained as a single locus in K. negevensis. Conversely, they split again into different loci in K. kingae. Subsequent to this split in K. kingae, a recombination event caused a duplication of the toxin genes in a single clade that is highly associated with invasive disease.
To determine whether duplication of rtxA results in more pathogenic strains, we constructed isogenic strains with zero, one, or two copies of rtxA and tolC. We also selected a panel of clinical isolates representative of both genotypes. We found that the isogenic two-copy strain secretes more RtxA and more rapidly traverses an epithelial barrier in vitro when compared to the isogenic one-copy strain. Importantly, RtxA is required for both of these assays, and the phenotypes are ablated in toxin-negative strains. However, when we extended these studies to our panel of clinical isolates, we observed no global trends between these two groups. Interestingly, the cytotoxicity and kinetics of epithelial barrier breach by the clinical isolates was highly variable, suggesting that strain background contributes significantly to toxin activity. Further study of this variation, and investigation into other systems that facilitate invasion by Kingella species is warranted.
Taken together, the results of this study reveal an uncommon mechanism of bacterial evolution wherein horizontally acquired genes require the co-option of existing systems for functionality. As the RtxA toxin appears to be required for invasive K. kingae disease, these are lynchpin events in the evolution of this important pediatric pathogen.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Great work. Very interesting. Thank you for sharing.