Breath-based COVID tests impacted by emerging variants
Published in Healthcare & Nursing
New technologies with ambitious approaches are being developed to screen for SARS-CoV-2, including breath tests. In fact, the US FDA approved the first breath test for COVID-19 under emergency use authorization (EUA) in April 2022.
Instead of measuring the virus directly, many breath-based tests for COVID-19 typically measure gas-phase metabolites we exhale that indicate our body is mounting a fight against the virus. Since late 2020, breath researchers around the globe have reported a "breathprint" of COVID-19 that identifies an infected individual.
In 2020, our team received funding to develop a breath test for COVID-19 from the United States National Institutes of Health (NIH) "RADx-rad" program, an initiative to support innovative approaches to address gaps in testing during the pandemic. As we were validating our test, we noticed our accuracy to identify infected persons began to decrease with the addition of samples collected after January 2022.
In our community, the primary circulating variant shifted from Delta to Omicron between December 2021 and January 2022. Since our models had been developed from samples collected from Delta-infected persons, we wondered if the decrease in our accuracy was the model struggling to identify an Omicron-specific breath profile.
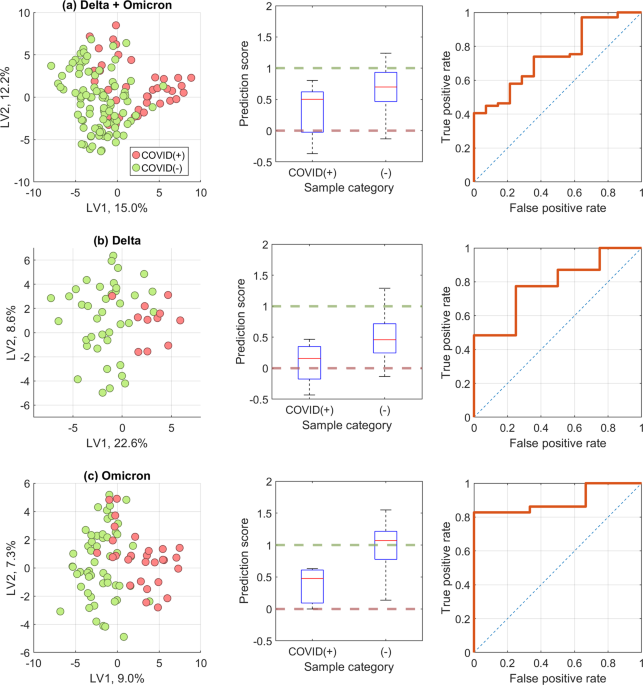
To test our hypothesis, we constructed three versions of our model. In one, we trained and validated a model using only breath samples collected during the Delta wave. In another, we only used samples collected during the Omicron wave. Finally, our third model was developed mixing samples of both variants, Delta and Omicron, together.
We found that modelling only one variant had a higher accuracy to correctly identify persons infected with that variant of SARS-CoV-2, rather than a universal model that attempted to model all data. Furthermore, there was a stark difference in the specific chemical contents of breath between Delta-infected and Omicron-infected persons. Models trained on Delta samples looked at 17 exhaled compounds to determine infection status, but the Omicron version required 35 compounds. Of these breath compounds, only ten overlapped between both model types.
Our findings were not surprising, given how different the presenting symptoms and clinical illness can be from these two variants. Delta is typically a lower airway infection that caused more serious disease, whereas Omicron usually infects the upper airway with symptoms typically milder than Delta. These differences in how our body responds to infection are reflected in our breath, which can complicate breath-based diagnostics.
We conclude that those developing breath-based assays for infectious airway pathogens such as COVID-19 should consider when dramatic viral variant or subtype shifts occur. Training models on a specific type might increase a diagnostic test accuracy from breath, as we determined with SARS-CoV-2.
Follow the Topic
-
Communications Medicine

A selective open access journal from Nature Portfolio publishing high-quality research, reviews and commentary across all clinical, translational, and public health research fields.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Healthy Aging
Publishing Model: Open Access
Deadline: Jun 01, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in