Characterization of an RNA binding protein interactome reveals a context-specific posttranscriptional landscape of MYC-amplified medulloblastoma
Published in Cancer
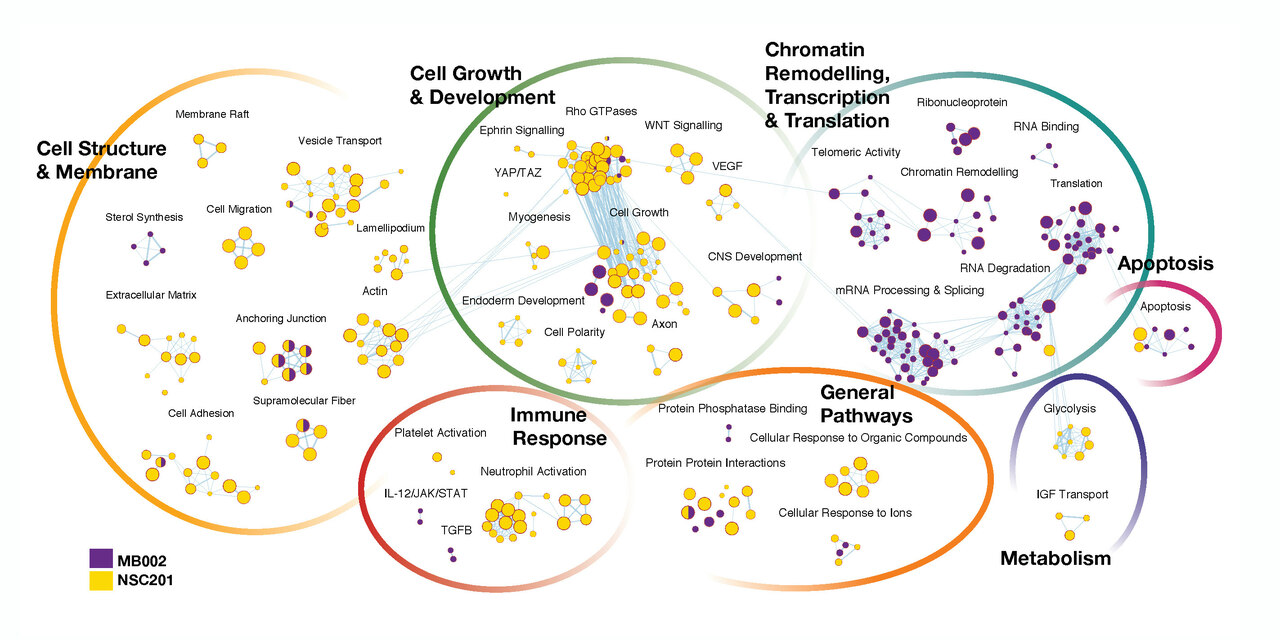
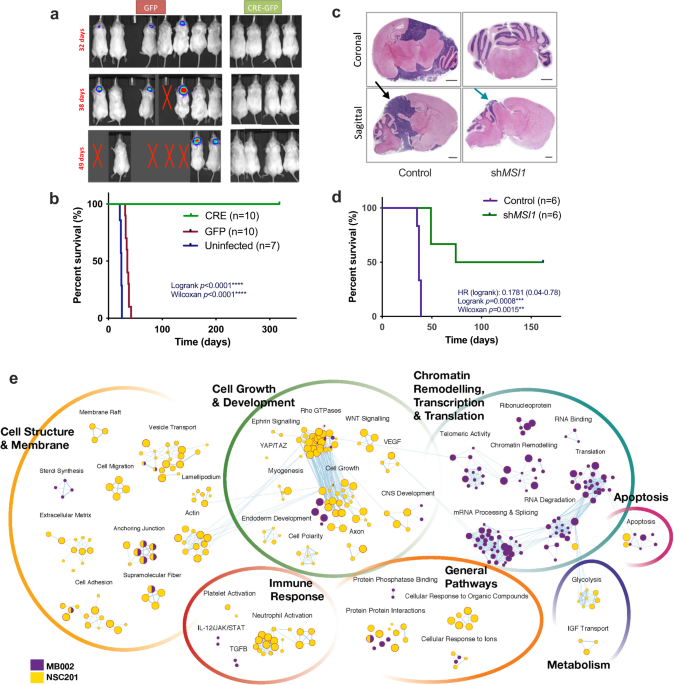
Recent advances in proteogenomics have suggested a requisite role of post-transcriptional modifications in G3 medulloblastoma (MB), including the MYC-amplified subtype (Group 3ã). Discordance between the transcriptome and proteome in G3 MB have suggested a central role of post-transcriptional gene regulation such as alternative splicing, alternative polyadenylation, RNA turnover and export. While genomic, epigenomic, and proteomic platforms continue to facilitate analyses of primary tumor tissue, the ability to directly link transcriptomic changes to the proteome remains a significant challenge. Overcoming these barriers is essential in studying neoplasms of the brain, which exhibits higher levels of post-transcriptional gene regulation required for neuronal development, plasticity and memory.
Musashi-1 (MSI1) is an RNA-binding protein (RBP) that maintains the multipotentiality of neural stem/progenitor cells during development, becoming rapidly downregulated in post-mitotic neurons. Similarly, cancer stem cells with high clonogenic potential in neural and non-neural tissues were found to highly express multiple neural stem cell determinant genes including MSI1. Since its discovery, multiple experiments have been designed to determine a functional role for MSI1 within the nervous system. Until recently however, biochemical protocols to reproducibly identify in vivo binding targets of an RBP with high fidelity limited any experimental conclusions.
Armed with the ability to directly study the in vitro binding partners of a RNA binding protein and to identify transcripts that are being actively translated, the possibility to directly measure the impact of a single RNA binding protein became feasible. We designed multiple large scale experiments to be later analysed using a mutiomic platform described by one of our senior authors.
The ability to maintain clinical utility of the multi-omic analysis was central to our research question. Not only did we want to identify a therapuetic target but also identify a neurodevelopmentally safer target for cancer therapy. In summary, we aimed to levereage the data available for neural stems cells and G3 MB in addition to generating our own new dataset to comparatively identify targets with the least neurodevelopmentally deleterious off target effects.
The positive energy and valuable input of the members of our international collaboration at each stage of this project was key from start to finish.

Open communcation between and interpretation of the multiomic analysis and the clinical relevance was the greatest challange and central to identifying a our target(s) for drug discovery.
I am so grateful to have had the opportunity to work with each an every contributor who offered their expertise throughout this project's journey.
Read the full article and let us know your thoughts of our approach to identifying a neural stem cell sparing, therapeutic target for MYC amplified G3 MB.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026
Latest Content
Why is Singapore Identified in Global Research as Number One? How Physical Activity and Education Excellence Created a Global Leader





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in