Charting biosynthetic diversity in big data
Published in Chemistry

Microbial biosynthetic pathways are an unimaginable source of chemical diversity, which is of great importance to our fundamental understanding of ecological interactions, as well as a key source of novel drug candidates for the development of, e.g., antibiotics and anti-cancer agents. When I started my research group at Wageningen University in 2015, one of my scientific goals had been to develop a computational framework to analyze biosynthetic pathways across large numbers of genomes and big (meta)genomic datasets. In previous years, during my PhD at the University of Groningen and my postdoc at the Max Planck Institute for Marine Microbiology in Bremen, I had (jointly with great collaborators) developed antiSMASH and MIBiG. Whereas the antiSMASH software made it possible to automate the identification of biosynthetic gene clusters (BGCs) in microbial genomes, the MIBiG data standard facilitated provided a reference database of BGCs of known function. This made it very convenient to analyze BGCs in individual genomes. However, it was clear that the field was moving towards massively scaled genome sequencing of thousands of microbes or diverse sets of microbial communities; this would clearly require new tools. After all, browsing through thousands of antiSMASH output HTML pages was obviously not an efficient way to analyze such datasets.
Interestingly, in the years before that, there had been parallel efforts to develop solutions for this. In one of these, I had been involved in myself, when I was a visiting scholar at the group of Michael Fischbach, who was at the University of California, San Francisco, in 2010-2011. In this study, we had developed a networking approach to map mutual similarities of large numbers of BGCs and group them into families. Simultaneously, the groups of Bill Metcalf and Neil Kelleher from Illinois had worked on a similar approach. A few months after I started in Wageningen in spring 2015, I co-organized a scientific meeting at the Joint Genome Institute in California, and Bill was one of the invited speakers. It was really nice meeting him, and in our discussions, we both realized that these first two studies were just the beginning, and that key challenges had to be overcome to democratize this technology. For example, the current implementations still required months of compute time on big servers or even supercomputers, to be able to take into account pairwise sequence similarity comparisons between thousands of BGCs. At that point, we decided to join forces.
A few months later, in the fall of 2015, a young and enthusiastic MSc student arrived in my group: Marley Yeong. He started working on an early implementation of what later became BiG-SCAPE, a tool that automated the calculation of sequence similarity networks directly from antiSMASH results and MIBiG reference gene clusters. In early 2016, Jorge Navarro-Muñoz arrived in Wageningen, who had recently moved into the field of bioinformatics and had received a grant of the Mexican Research Council (CONACYT) to visit my lab for 12 months as a postdoc. Under his leadership, the BiG-SCAPE project evolved into a large software project, with various contributions from students within the group and collaborators outside: for example, my former colleague Antonio Fernandez-Guerra from the MPI in Bremen contributed to selection of clustering algorithms; Emzo de Los Santos, whom Jorge had met when he presented his work at an NPRONET meeting in the UK, contributed to various parts of the code base, Sahar Abubucker from Novartis came over for a sabbatical to develop a prototype for an HTML visualization of the BiG-SCAPE output, and Satria Kautsar, who arrived as a PhD student in my group, developed a full-fledged JavaScript-enabled visualization, through his software engineering expertise.
When analysing the first results of BiG-SCAPE, it also became clear that incomplete clusters could have a large impact in the pairwise distance measurement, so we started to think about strategies to cope with this. This led to the development of BiG-SCAPE's glocal mode (and the annotation by antiSMASH of partial clusters with the 'contig_edge' flag). At the same time, several new computational innovations were introduced that made the whole pipeline much faster.
In 2017, I visited the International Symposium on the Biology of Actinomycetes in Korea, and met with Francisco (Paco) Barona-Gomez and his team. Nelly, who was a PhD student with Paco, had been developing a really nice tool, named CORASON, which provided a strongly complementary approach to our efforts on BiG-SCAPE, by facilitating the large-scale phylogenomic analysis of BGCs across large sets of genomes, based on multi-locus phylogenies of the common core shared by these gene clusters. Nelly's visit to Wageningen at the end of 2017 was crucial to integrate the CORASON approach into the last stage of BiG-SCAPE, and to define the outline of the paper, presenting how both tools could work together to analyze biosynthetic diversity across large genomic datasets.
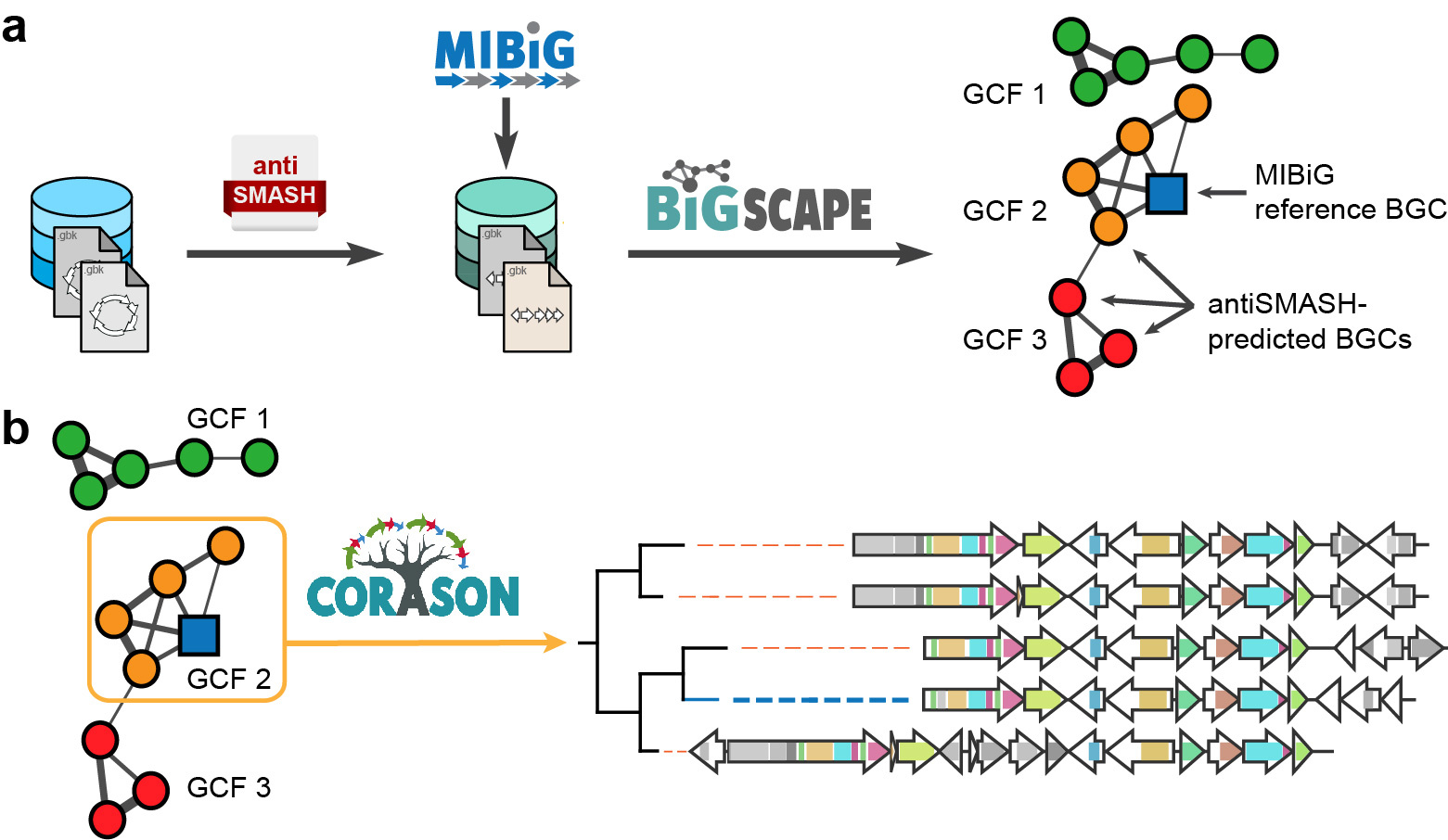
The BiG-SCAPE / CORASON workflow uses sequence similarity networking to group biosynthetic gene clusters into families and applies multi-locus phylogenetic analysis to identify their evolutionary relationships.
Within this same year, videoconferencing between our group and those of Bill Metcalf, Neil Kelleher and Regan Thomson, intensified. The Illinois and Northwestern groups had been following up on their initial observations by developing a platform known as metabologenomics, which links biosynthetic gene cluster families to the metabolites they produce by combining the output from bioinformatics tools similar to BiG-SCAPE with mass-spectrometric metabolic data. Michael Mullowney, who had started a postdoc with Neil and Regan, happened to be brainstorming at that time about the potential of exploring the unusually large and diverse detoxin/rimosamide class as a project. This turned out to be a fantastic case study to showcase how the BiG-SCAPE / CORASON tandem could be applied to systematically map chemical diversity across a large set of related BGCs. After a lot of hard work, Michael was able to isolate detoxins with diverse chemical modifications which corresponded to the genetic diversity that had been computationally predicted. At the same time, James (Hudson) Tryon, a PhD student in Neil’s lab, had been able to confirm that sensible metabologenomic correlations could be made with gene cluster families from BiG-SCAPE to correlate these to the presence of molecules in metabolome data from a large set of actinomycete bacteria.
What followed in the ensuing years was a lot of ‘dotting i’s’ and ‘crossing t’s’, and real team efforts by all authors to turn this into a round story that was easy to follow and illustrated by esthetically pleasing figures. In the end, a paper like this was really only possible by combining all of our expertise, and I feel that the type of collaborative spirit that we shared is really what drives scientific progress. I, and the whole team with me, hope that the new tools we introduce will be very useful to the research fields of natural product discovery and microbiome ecology, and look forward to interacting with the community to help to put them to good use and improve them further.
The Streptomyces spectabilis NRRL 2792 strain that produces detoxins N1-3 shows a beautiful pink color in the lab, as it also produces prodiginins. Picture credit: Michael Mullowney.
Credit for poster image on top: Michael Mullowney.
Check out Big-SCAPE and CORASON here.
Follow the Topic
-
Nature Chemical Biology

An international monthly journal that provides a high-visibility forum for the chemical biology community, combining the scientific ideas and approaches of chemistry, biology and allied disciplines to understand and manipulate biological systems with molecular precision.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
We used BiG-SCAPE / CORASON for the analysis of a gene cluster among some fungal species, and I am glad to hear the story behind it. That was really interesting for me. Best wishes to you and your team.