Evolution can repeat itself and instances of parallel evolution in response to similar environmental pressures can provide evidence of evolution by natural selection. On the other hand, idiosyncratic outcomes can be driven by chance or contingency in evolution and indicate constraints on the power of selection in driving evolution. Several studies have indicated striking parallelism at the genomic level which is often driven by single genes of major effect such as the Ectodysplasin gene controlling body armor has repeatedly been used by numerous populations of stickleback fish during freshwater adaptation. Likewise, the Agouti and Mc1R genes control coloration in diverse organisms. While repeated evolution driven by single genes has been documented well, parallelism is less understood when evolution is driven by many genes of smaller effect with only a few studies showing genome-wide variation. Moreover, evolution is not always parallel, and the probability and extent of parallelism can decline as the time of divergence increases between taxa. While studies have documented this decline in parallelism, few have dissected the potential complex causes that underlie this decline.
My interest in studying the predictability of evolution developed during my Ph.D. and it was rather timely that Dr. Patrik Nosil was visiting my Ph.D. advisor Dr. Zach Gompert’s lab on a sabbatical at Utah State University. After a quick chat with Patrik about geographical variation in Timema, I expressed my interest in working on this project. This started as a side project and was four years in the making only to teach me that good scientific stories take time to develop and publish. Defying Patrik’s belief that side projects do not finish on time and spanning three jobs for me, here we are with this interesting piece of work that adds to the growing literature on climate change genomics.
In this paper, we investigated the genomic basis of parallel adaptation to a novel ecological dimension i.e., climate in eight species of wingless, univoltine, herbivorous stick insects in the genus Timema (Figure 1A) most of which are endemic to California, USA. Within their geographic range, Timema occupy variable habitats which range from sea-level to mountainous regions, and from arid semi-deserts near the Mexican border to wet evergreen forests in northern California. This variation in habitat and previous evidence of parallel evolution provides an ideal and powerful background to study climate adaptation in these insects. Moreover, a species-level sampling of organisms across their geographic range is a rare advantage that provides immense power to study the decay of parallelism. For this study, we tested the shared ecology and genetics hypothesis in Timema to identify climate-associated gene regions within species which show a range of divergence times of up to tens of millions of years (here, generations). We assessed the contribution of shared ecology and genetics to genomic parallelism by comparing the proportions of the genome that exhibit repeated genotype-climate association and identified modest decay in parallelism with increased ecological and genetic distance for a given pair of species. We then bolstered the evidence that climate-associated gene regions are likely subject to selection by using a field experiment and genetic mapping of cuticular hydrocarbons. Our collective results yield a comprehensive evaluation of genome-wide parallel evolution in the context of an environmental pressure of high current interest (i.e., climate), and in a system where comparison can be made to parallelism seen at a single, major locus (i.e., Mel-Stripe). In this blog, we highlight two crucial points from our paper – first, we discuss our results for the shared ecology and shared genetics hypothesis, and second, we highlight the importance of cross-validation of tests for genomic parallelism which are important as we tackle the several inherent limitations of genomic data.
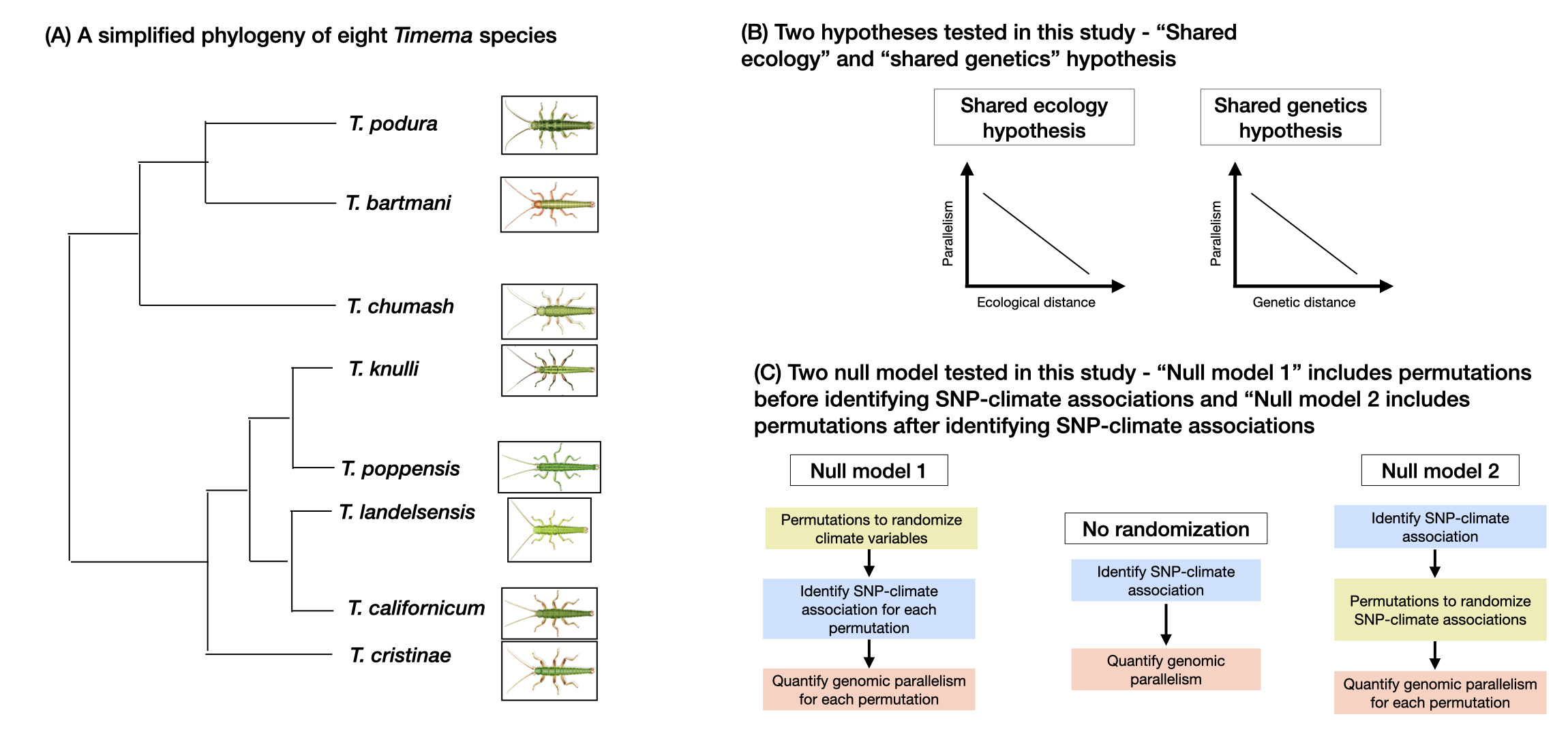
Figure 1: (A) A simplified phylogeny of eight Timema species included in this study. (B) Two hypotheses that we use to test for the decay of parallelism: The first diagram shows our prediction for the “shared ecology” hypothesis where we expect a decay in parallelism with an increase in climate (i.e., habitat and ecological) distance. The second diagram shows our prediction for the “shared genetics” hypothesis where we expect a decay in parallelism with an increase in genetic distance. We use these two hypotheses to study the decay of parallelism. (C) Our two null models which we use to test the robustness of parallelism as detected by our statistical tests. The first flow chart depicts a workflow of the first null model ("Null model 1") where the climate (Principal components) variables were randomized before identifying SNP-climate associations. This test was implemented for all eight species and 56 species pairs. Estimates of parallelism for each permutation were then compared to estimates of parallelism from the second flowchart where no randomization was performed. The third flowchart depicts a workflow of the second null model ("Null model 2") where the permutations were performed after identifying the SNP-climate association. This test was implemented for all eight species and 56 species pairs. Estimates of parallelism for each permutation were then compared to estimates of parallelism from the second flowchart where no randomization was performed.
What does the “shared ecology” and “shared genetics” hypothesis tell us about decay in genomic parallelism?
While the decline of parallelism in relation to time since divergence is well established, the potential causes that affect this decline are less understood. In this context, we tested two general hypotheses which we refer to as the "shared ecology" and "shared genetics" hypotheses (Figure 1b). Under the shared ecology hypothesis, we expected that parallel evolution is more likely when shared ecologies result in similar patterns of natural selection in different taxa such as ecotypes or divergent lineages. Shared aspects of environmental variation can decline with time since divergence, as species (or even populations or ecotypes) come to occupy different geographic areas or as local environments change over time, thus reducing parallelism at both phenotypic and genotypic levels. Likewise, under the shared genetics hypothesis we expected parallelism to be more likely when genomes are similar because of pools of standing variation, new mutations which arise, and the effects of these mutations will tend to be more similar in closely related genomes. Both ecological (i.e., habitat and climatic) and genetic similarity are expected to decline with time and there is support for both hypotheses. Our results revealed evidence for the effects of both ecology and genes on the extent of genomic parallelism with some variation based on the climatic variable considered. These results suggested that observed genomic parallelism in this system can be partly attributed to selection pressures exerted on these insects as they inhabit similar climatic niches. Moreover, these results agreed with other studies and indicate that similar gene modules which possibly have the same functional role are driving convergent adaptation to climate.
Why do we need to cross-validate tests of genomic parallelism?
The choice of an appropriate null model is fundamental in validating statistical inferences about genomic parallelism. Different null models can be sensitive to different aspects of the ecological and genomic data and can therefore lead to false positive signals of parallelism. To account for hidden biases in our data, we conducted two sets of permutation tests where our first null model (Figure 1c – Null model 1) included randomization of climatic variables before identifying single nucleotide polymorshism (SNPs) association with climate (hereafter called “SNP-climate associations”). For our second null model ((Figure 1c – Null model 2), we first identified SNP-climate associations and then performed permutations to randomize these associations to identify parallelism for each permutation as compared to our original data. We did so to ask whether the patterns of observed genomic parallelism and its decay could have been inflated by unaccounted aspects of the genetic data, such as shared SNP density in specific genomic regions, allele frequency distributions, or linkage disequilibrium, affecting some genomic regions more than others. We found no evidence of the observed decay in parallelism with climatic or genome-wide divergence in permuted data sets conducted prior to or following the identification of SNP-climate association. Overall, these findings in combination with the experiment and CHC results provide the support that the documented parallelism in genomic association with climate reflects a contribution from the selection. However, we also note that our analyses using permuted data sets generated instances where ‘significant’ x-fold excesses in the numbers of gene regions displaying parallelism above null expectations. This approach involving permuted data sets highlights important issues concerning the analytical aspects of parallelism tests. Our findings thus concur with previous studies using simulation-based approaches showing that false positives can be detected due to unaccounted aspects of the genetic data. Therefore, we suggest that these associations should be interpreted with caution, and studies identifying genomic association with climatic variables warrant additional cross-validation of findings, as performed here.
Why is this study important for studying climate adaptation?
While studies on the repeatability of evolution are crucial for understanding the evolution of biodiversity, they also offer an interesting insight into how organisms will adapt to changing climates over time. Our study helps in our preliminary effort to understand the genomic basis of adaptation to climate by highlighting that in this case adaptation is driven by a complex genetic architecture and is not driven by a single genomic region or single mutation. Instead, several regions of the genome underlie adaptation to climate and are possibly several phenotypes that interact to drive this adaptive scenario. This sets up an ideal premise for future studies to predict species-level response to climate by using genomic vulnerability models and use functional genomics to validate phenotypes associated with key climatic variables such as temperature, elevation, and precipitation. Finally, our study highlights the importance of geographic sampling of wild organisms over a large phylogenetic and spatial scale which can provide a powerful dataset to answer questions related to ecological dimensions both at phenotypic and genomic levels.
This project highlights the combined patience of all the coauthors and the endless discussions with colleagues who have been inspired by this project to pursue similar research in other systems.
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026
Latest Content
Why is Singapore Identified in Global Research as Number One? How Physical Activity and Education Excellence Created a Global Leader





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in