Combinatorial single-cell profiling of major chromatin types with MAbID
Published in Protocols & Methods

Deciphering gene expression is a complex task, involving the study of coordinated regulatory mechanisms across different genomic scales. While numerous single-cell sequencing technologies focus on individual aspects of gene regulation, such as mRNA levels, histone modifications, DNA methylation, or chromatin accessibility, methods capable of measuring multiple layers of gene regulation within the same cell are scarce. Some existing approaches can study two to three factors simultaneously, but they predominantly rely on Tn5-based tagmentation and primarily focus on active chromatin and facultative heterochromatin types. Recognizing this gap, we sought to develop an alternative method to comprehensively investigate all chromatin types with multiplexed measurements at the single-cell level.
As members of the Kind lab at the Hubrecht Institute - led by Jop Kind, a professor by special appointment in Single Cell Epigenomics at Radboud University Nijmegen and an Oncode Investigator - we are dedicated to unraveling the principles governing cellular decision-making, specifically in the acquisition of new identities and traits. By developing techniques such as scDam&T-seq, EpiDamID, and now MAbID, we explore the role of genome architecture and chromatin context in the temporal and spatial control of gene expression during development and disease. For more details about our lab, please visit our Hubrecht Institute and Oncode Institute websites.
The development of MAbID was an exciting journey for our team. Our aim was to create a method that allows researchers to jointly profile multiple epigenetic markers in a single sample. We based our design on antibody-DNA conjugates, in which each antibody is covalently conjugated to a uniquely-barcoded DNA adapter. Firstly, the genome is digested using standard restriction enzymes, which create a compatible handle for ligation of the antibody-specific adapter. This antibody-adapter ligates at the genomic binding site of the antibody, thereby acting as a proxy for the epigenetic mark's localization. Moreover, the unique barcode in the adapter enables us to classify which modification was measured, facilitating the multiplexing of several measurements in the same sample.
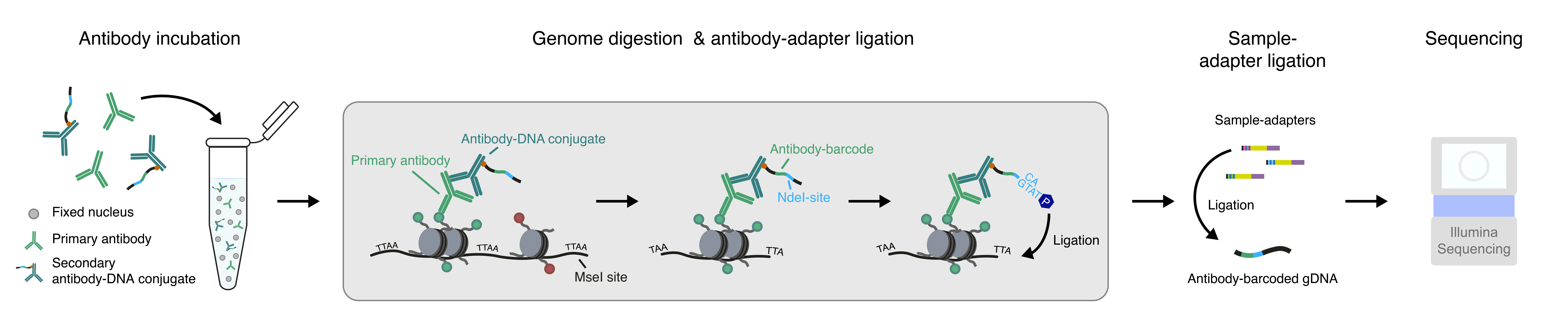
Our initial results were promising, providing specific genomic profiles for the Lamin B1 protein in the first attempt. Over the years (and a couple of name changes, aided by polls on our group’s Slack channel) we continually advanced both the experimental and computational aspects, which was very exciting. For example, we realized that by including another restriction enzyme besides our original MseI, we could potentially increase the resolution of the method and tailor the protocol to the protein-DNA interactions of interest. We were very happy to see that by also targeting MboI-sites, we could indeed increase the quality of the data.
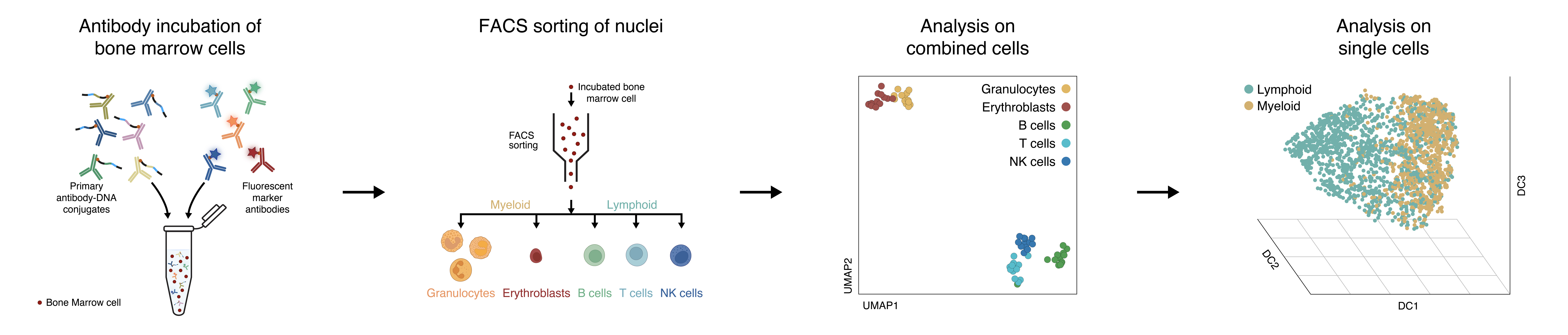
Then we moved towards generating MAbID data from single cells. We started in mouse embryonic stem cells cells with six targets, which worked so well that we decided to use this approach to study in vitro neural differentiation towards neural progenitor cells. This six-epitope single-cell dataset of multiple cell states was a great resource to computationally dig into. We found that we could identify cells undergoing X-Chromosome Inactivation and capture the accompanying changes in epigenetic markers on the inactivated X-allele. Next, we turned our attention to using scMAbID in primary tissues, since this implementation could make the method more widely applicable to various research questions. For this, we sorted five different mouse bone-marrow cell types and used it to show that the data holds the lineage- and cell-specific epigenomic information.
We are excited to have developed MAbID and hope it will benefit others to generate multiple epitope-binding tracks from their preferred systems, whether with limited or abundant cell numbers. We believe it holds great potential for future applications, such as studying the combined epigenetic landscape of complex biological systems. This approach could deepen our understanding of the coordination between gene-regulatory mechanisms and the single-cell resolution can shed more light on the cellular heterogeneity. We are eager to see how MAbID will be used in developmental systems and clinical settings.
For more information, you can find the paper in Nature Methods, and the bioinformatics tools are available on Github.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in