Commensal to commander – the global spread of multidrug-resistant Staphylococcus epidermidis
Published in Microbiology

The paper in Nature Microbiology is here: https://go.nature.com/2QczSRW
Advances in modern medicine mean remarkable things can be done to save people, but many of these are invasive. Each time the skin is breached and something foreign placed in the body (be it simple like a cannula into a vein to give medicines, or complex like a joint or heart valve replacement) this creates an opportunity for S. epidermidis to enter and create a focus of infection. As a result, S. epidermidis is one of the most common causes of hospital related infections. Many of these infections occur in the Intensive Care Unit where patients are sickest and often immunocompromised, with catheters inserted into their bodies to deliver treatments.
Vancomycin is a last-line agent in the treatment of staphylococci, but in these times of increasing antibiotic resistance, it is routinely used first-line for the treatment of S. epidermidis. In serious and difficult to treat infections, the combination of vancomycin plus rifampicin is used, based on the assumption that these two unrelated antibiotics should protect one another against developing resistance to both. But what happens when the “conventional wisdom” on which we base treatment decisions may be incorrect?

Macromethod Etest screening for reduced glycopeptide (vancomycin and teicoplanin) susceptibility. Image Jean Lee.
After noticing an increasing number of difficult to treat S. epidermis clinical infections (that were resistant to rifampicin and failed to respond to vancomycin) were occurring in tertiary hospitals around Melbourne, Australia, we wondered if the strains causing these infections were related. What followed evolved into an international collaborative study in which whole genome sequencing was used to reveal that three lineages of near pandrug-resistant S. epidermidis have been spreading through global hospitals over the past 17 years, without recognition that they were clonally related.
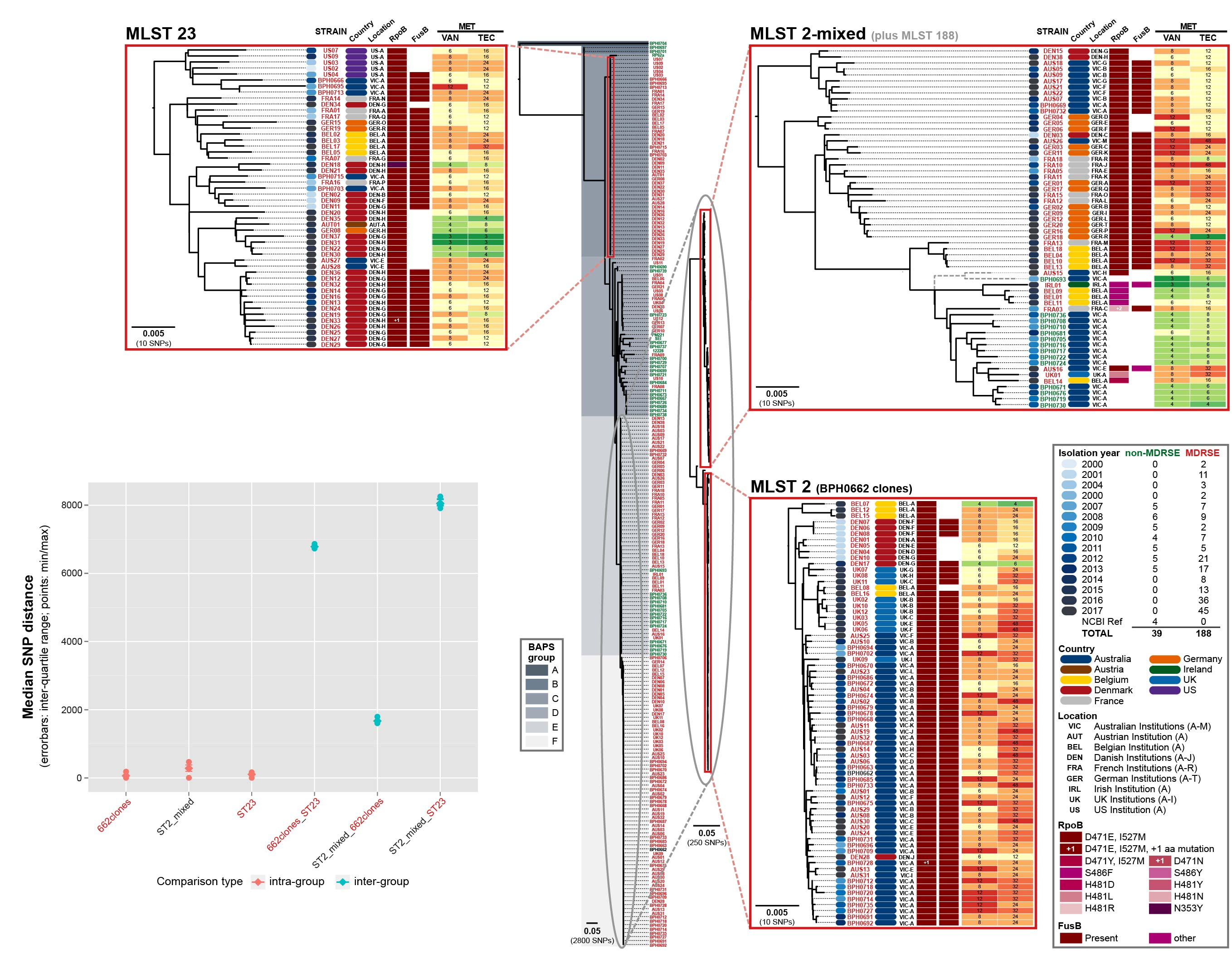
Three multidrug-resistant lineages of S. epidermidis sharing a dual D471E and I527M RpoB substitution have spread globally.
A dual substitution (D471E and I527M) in RpoB that causes high-level rifampicin resistance was identified as common to clones in the three lineages. Through allelic exchange, we were able to demonstrate that this dual RpoB mutation also caused cross-resistance to vancomycin. This suggests that combined use of vancomycin and rifampicin has likely contributed to increasing resistance to both these agents rather than prevention as intended. With standard treatments exhausted, only salvage drugs (for which evidence about clinical outcomes are limited and side-effects common) remain. Alarmingly, some of the European isolates in our study have also acquired resistance to the salvage antibiotics, rendering them potentially untreatable.
The new recognition of these drug-resistant S. epidermidis lineages raises many questions that will need to be addressed in future studies: Why is this problem occurring in a time when infection prevention and control practices have reduced hospital MRSA?; What is the true clinical impact of infection with these strains?; What is the ideal approach to therapy?
Written by Jean YH Lee and Benjamin P Howden

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in