Distance-AF improves predicted protein structure models by AlphaFold2 with user-specified distance constraints
Published in Protocols & Methods, Cell & Molecular Biology, and Mathematics
Protein structures are essential for understanding biological functions and advancing drug discovery. While AlphaFold2 (AF2) revolutionized protein structure prediction with near-experimental accuracy, it still faces key challenges: it often mispositions multi-domain proteins, produces incorrect elongated loops, and predicts only single static structures despite many proteins adopting multiple functional conformations. To address those limitations, we developed a new approach that builds on AF2 by integrating user-defined distance constraints between amino acids. This allows researchers to guide structure prediction using experimental constraints.
How Distance-AF works
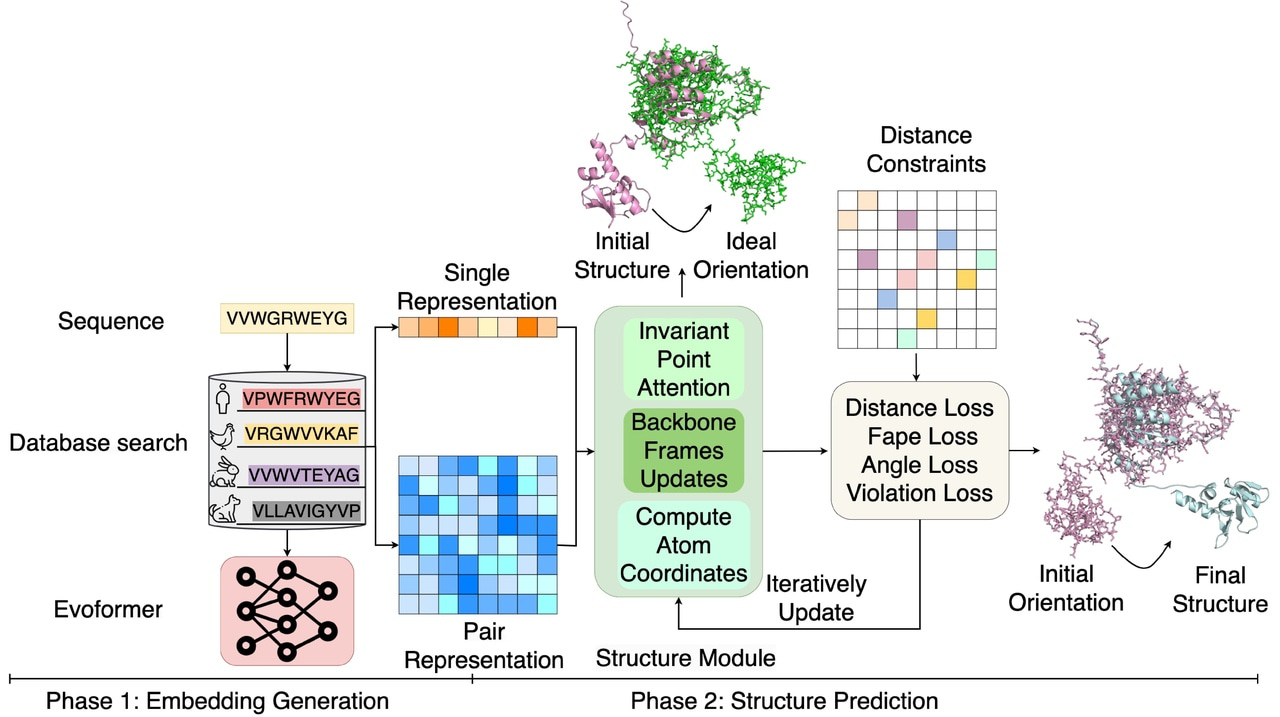
Distance-AF consists of two main components (Figure 1 a), 1). processes multiple sequence alignments into embeddings, identical to AF2. 2) Constraint-Guided Structure Module: Takes user-defined inter-residue distance constraints and iteratively refines the structure by minimizing a distance loss function alongside AF2's standard losses. In Figure 1 a, we show the overall architecture. The sequence goes through database search and the Evoformer to generate pair representations. These feed into the Structure Module along with user-specified distance constraints. The module iteratively updates through its Invariant Point Attention Backbone, incorporating multiple loss functions (Distance Loss, Fape Loss, Angle Loss, Violation Loss) until the predicted structure satisfies the constraints.
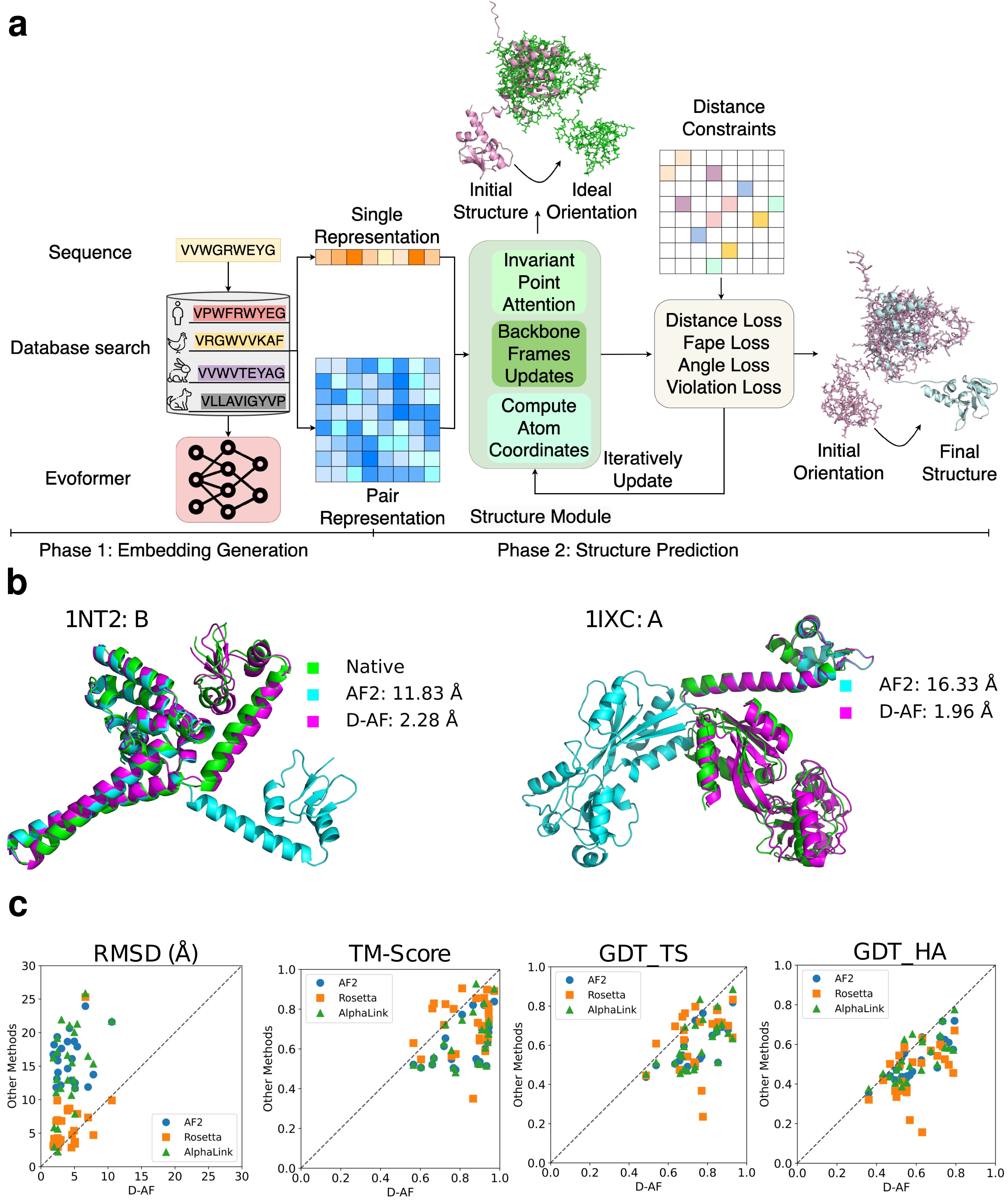
Figure 1. Distance-AF workflow and benchmarking results. (a) Distance-AF architecture showing two main phases: Phase 1 (Embedding Generation) and Phase 2 (Structure Prediction). (b) Representative examples comparing Distance-AF (D-AF, magenta) predictions to vanilla AlphaFold2 (AF2, cyan) and native structures (green). (c) Benchmark performance across 25 non-redundant protein targets comparing Distance-AF, vanilla AF2, Rosetta and AlphaLink across four metrics: RMSD, TM-Score, GDT_TS, and GDT_HA.
Performance Results
Figure 1 b illustrates two examples which are accurately predicted by Distance-AF while AF2 struggles on predicting the correct domain orientation. Distance-AF (D-AF, magenta) dramatically improves over AF2 (cyan) when compared to native structures (green). For instance, in example 1NT2: B(left panel of Figure 1 b), Distance-AF achieved 2.28 Å of RMSD which AF2 has 11.83 Å, and in example 1IXC: A(right panel of Figure 1 b), Distance-AF reduced the RMSD from 16.33 Å to just 1.96 Å by correctly positioning the domains.
Figure 1 c shows the benchmarking results of Distance-AF with other methods. We benchmarked Distance-AF against vanilla AF2, Rosetta, AlphaLink on 25 non-redundant targets. Distance-AF achieved an average TM-Score of 0.834 (with six distance constraints), substantially outperforming AF2 (0.622), Rosetta (0.728), AlphaLink (0.644).
Distance-AF also excelled across diverse applications, we further demonstrated in the paper that it reduces average RMSD from 9.47 Å to 3.16 Å for cryo-EM structures and from 9.53 Å to 2.34 Å for proteins with flexible linkers.
Conclusion
Distance-AF effectively incorporates experimental or predicted distance information to improve protein structure prediction, particularly for multi-domain proteins and flexible regions. We successfully applied Distance-AF to various challenging scenarios including cryo-EM structures, NMR targets, disordered loop region targets, and GPCRs.
The method is most effective for structured domains requiring repositioning and proves robust to moderate constraint perturbations (up to 5.0 Å noise).
The full source code and tutorials are available at:
GitHub: https://github.com/kiharalab/Distance-AF
Zenodo: https://zenodo.org/records/16891488
Tutorial: https://github.com/kiharalab/DistanceAF/blob/main/README.md
For questions or suggestions, contact Prof. Daisuke Kihara at dkihara@purdue.edu.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in