Connecting AI & Pathology Using TIAToolbox
Published in Bioengineering & Biotechnology

During surgery or biopsy, suspicious or diseased tissue is removed, fixed in paraffin or frozen before being sliced into very thin tissue sections which are stained, placed on glass slides and examined under a microscope by a pathologist. The pathologist then advises clinicians on their diagnosis, crucial for selection of appropriate treatment and prognostication if needed. Over the last 5 years, this process has been transitioning to the use of digital slide scanners which produce a high-resolution Whole Slide Image (WSI), typically consisting of billions of pixels for each slide. Among other benefits for technology in education and seamless requesting of second or third opinions, digitization of pathology has also contributed to 10–15% gain in reporting via streamlined diagnostic workflows1.
The number of cases and sections to analyze have increased over the years, which, coupled with varying degrees of case complexity, has resulted in a significant increase in workload of an already stretched pathologist workforce2. According to the latest Royal College of Pathology workforce survey, only 3% of NHS hospitals in the UK report adequate staffing. Therefore, there is a need to automate the analysis of WSIs to aid the pathologists in performing diagnosis. In our recent work3, we report the development of the Tissue Image Analysis Toolbox (TIAToolbox) that will help standardize and accelerate the development of such automated analysis pipelines by providing a wide range of functionalities — from reading whole slide images to training artificial intelligence (AI) models.
It is common to ‘reinvent the wheel’ or write monolithic use-case specific code with inappropriate quality checks in place4 when implementing Computational Pathology (CPath) pipelines. A major aim of TIAToolbox is to make it easy for researchers to reuse and adapt existing pipelines. TIAToolbox is built from sturdy and reliable components, with each component having clearly specified inputs and outputs. It uses a modular design to reduce code complexity which makes the code easier to understand and maintain. Furthermore, it enables advanced users to easily modify or replace a component.
The toolbox allows for development of complex WSI image analyses by providing robust support for simple tasks such as feeding images to downstream analysis using pre-trained deep learning methods. It provides support for five major components of whole slide image analysis pipelines: data loading, pre-processing, tile level or localized tissue analysis, whole slide image level predictive modeling and visualization.
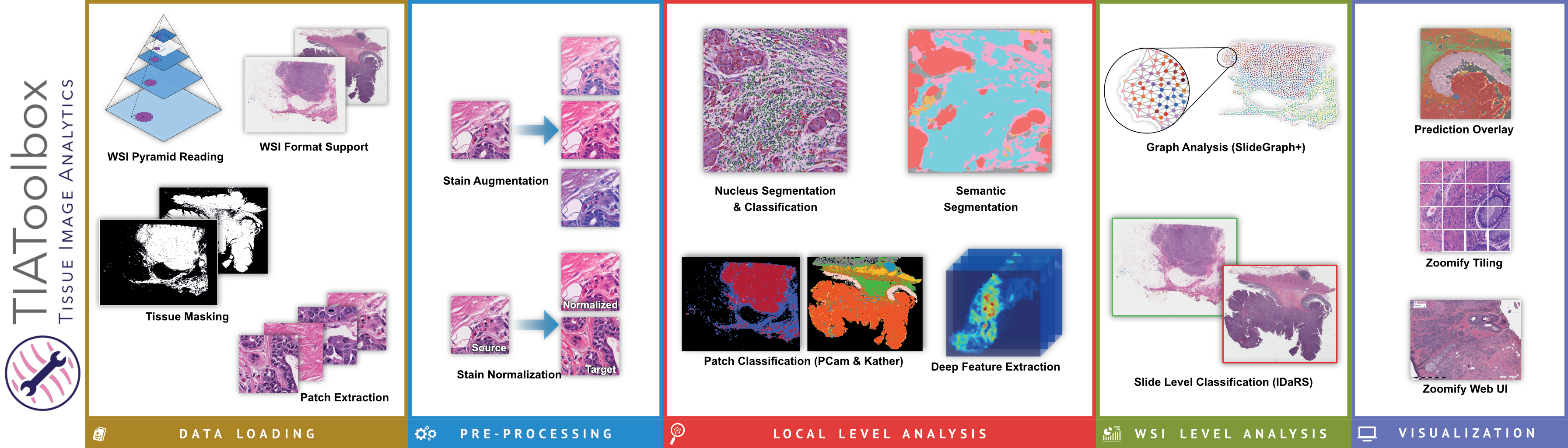
One of the most common tasks when constructing a CPath pipeline is simply reading pixel data from a WSI file. These large multi-gigapixel image files can be challenging to handle, often requiring specialized tools. Additionally, there are many file formats currently in use, requiring different software libraries for decoding data, where each of these libraries have their own interface. When working with data from multiple centers and in multiple file formats, it can be very challenging to write code which is compatible with these multiple formats. Furthermore, the images may have been scanned at different resolutions. For many methods, the tissue image must be normalized to be of the same resolution. While this information is embedded in the WSI file, it must be extracted and used to resample the image data. This can again be a different process across formats. We provide WSI readers in TIAToolbox which ensures a consistent interface when working with many file formats. This can greatly simplify the code required for working with multiple file formats.
Another example of a common CPath task is cell nucleus detection or segmentation. For instance, you may wish to segment all cells within an image before performing some analysis of the variation in the shape and size of cells between malignant and benign tissue samples. Accurate nucleus detection and segmentation are very challenging tasks, currently best solved by a deep learning model such as a convolutional neural network (CNN). However, training a model for such a task is a significant undertaking and can be a barrier to exploration and experimentation of methods. By including the code for running nucleus detection and segmentation model inference in addition to pre-trained weights, we hope to make this more accessible to new researchers in the field and enable easier pipeline development for experimentation for everyone in the field.
The output of a step such as nucleus segmentation may result in several million cell boundary polygons. Handling this volume of polygon annotation data can be quite challenging, not only in terms of creating a storage efficient representation on disk but also for fast and resource efficient querying. We provide tools to handle this kind of output to efficiently process several million polygon annotations and execute performant queries. Typical spatial queries with our toolbox can be performed in under one tenth of a second (see the benchmark notebook on GitHub), even with several million polygons in the database.
We include modules for many other tasks performed as part of a typical CPath pipeline, including but not limited to stain separation, stain normalization, tissue classification, and weakly supervised learning, and graph based whole slide image level predictive modeling. We hope to continue to expand this feature set over time in addition to continuing to maintain or improve the code quality, test coverage, and documentation.
By making these tools available under a permissive BSD license (and model weights under creative commons licenses), we invite other researchers and developers to contribute and hope that TIAToolbox will be a valuable asset to the CPath community. Additionally, the commercial-friendly licensing enables TIAToolbox to help accelerate developments in computational pathology, for which the market is expected to grow by 7.5% compound growth annually5 (78% in total) from 2022 through 2030.
We are pleased to see rapid adoption of TIAToolbox, which has been downloaded over 111,000 times, by other researchers in the field6–9. TIAToolbox has additionally been used as part of several entries to CPath challenges, including the CoNIC challenge9.
Full documentation for TIAToolbox, including code snippets and full pipeline implementation examples, can be found on Read The Docs at https://tia-toolbox.readthedocs.io. Additionally, the full source code can be found on GitHub at https://github.com/TissueImageAnalytics/tiatoolbox.
References
- Martin, J. Developing a digital future for pathology. 8th Emirates Pathology & Digital Pathology Utilitarian Conference. Online at https://www.youtube.com/watch?v=DAxoiFtGvBA&t=145s (2021).
- The Royal College of Pathologists. Meeting pathology demand: Histopathology workforce census. Online at https://www.rcpath.org/uploads/assets/952a934d-2ec3-48c9-a8e6e00fcdca700f/Meeting-Pathology-Demand-Histopathology-Workforce-Census-2018.pdf 3 (2018).
- Pocock, J., Graham, S., Vu, Q.D. et al. TIAToolbox as an end-to-end library for advanced tissue image analytics. Commun Med 2, 120 (2022). https://doi.org/10.1038/s43856-022-00186-5
- Singh Chawla, D. (2020). Critiqued coronavirus simulation gets thumbs up from code-checking efforts. Nature, 582, 323–324. https://doi.org/10.1038/D41586-020-01685-Y
- Grand View Research. Digital Pathology Market Size, Share & Trends Analysis Report By Application (Academic Research, Disease Diagnosis), By Product (Software, Device), By End-use (Diagnostic Labs, Hospitals), And Segment Forecasts, 2022 - 2030. (2022).
- Lu, Wenqi et al. "SlideGraph+: Whole Slide Image Level Graphs to Predict HER2 Status in Breast Cancer." Medical Image Analysis (2022): 102486. https://doi.org/10.1016/j.media.2022.102486
- Shmatko, A., Ghaffari Laleh, N., Gerstung, M. et al. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer 3, 1026–1038 (2022). https://doi.org/10.1038/s43018-022-00436-4
- Hameed, Z., Garcia-Zapirain, B., Aguirre, J.J. et al. Multiclass classification of breast cancer histopathology images using multilevel features of deep convolutional neural network. Sci Rep 12, 15600 (2022). https://doi.org/10.1038/s41598-022-19278-2
- Graham, S. et al. CoNIC: Colon Nuclei Identification and Counting Challenge 2022. Preprint at https://doi.org/10.48550/arXiv.2111.14485 (2021).
Follow the Topic
-
Communications Medicine

A selective open access journal from Nature Portfolio publishing high-quality research, reviews and commentary across all clinical, translational, and public health research fields.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Healthy Aging
Publishing Model: Open Access
Deadline: Jun 01, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in