Controlling drug resistance with evolutionary principles
Published in Cancer
Evolution is a process of random mutation and non-random selection. In cancer, evolution drives treatment resistance, arguably the biggest problem in cancer therapy today (Greaves & Maley, 2012). Evolution, both in species and in cancer, is unstoppable.
If cancer evolution is unstoppable, is it at least predictable?
The contingencies of evolution are numerous, often leading to stochastic outcomes (Gould, 1989). However, sometimes evolution seems predictable, for example where it leads to convergent phenotypes, such as fins in fish and cetaceans. Given enough standing variation and strong selective pressure, evolution may be highly predictable. In cancer we have >1010 genomically unstable cells and strong selective pressures from treatments that leads to convergent resistant mechanisms (McGranahan & Swanton, 2015).
If cancer evolution is predictable, can we control it?
Collateral drug sensitivity is an established concept in oncology (Hutchinson, 1963; Pluchino et al. 2012; Zhao et al. 2016). If we first prescribe drug A, and resistance then evolves, is the resistant tumour more susceptible to a follow-up drug B? Our interest in this phenomenon and its clinical implications originated from a collaboration with Prof Udai Banerji from the Cancer Research UK Cancer Therapeutic Unit at the ICR. In the clinic, using drugs in sequences rather than in combination can significantly reduce toxicity.
Collateral drug sensitivity can have an evolutionary explanation, as resistance can incur a cost when the environment changes. Such evolutionary trade-offs can be exploited clinically through ‘evolutionary steering’ (Nichol et a. 2015), wherein drugs are used to drive evolution in our favour.
The difficulty is to know not just what drugs to use, but also when to switch drug. This may also be patient-specific, requiring to monitor cancer evolution over time and use evolutionary theory to inform on the next treatment decision.
We wanted to test evolutionary steering experimentally. Our lab (www.sottorivalab.org) is a mixture of computational and experimental cancer research, so we put together a molecular biologist (Ahmet Acar) and a computational biologist (Daniel Nichol) to work on the problem. It proved to be a fantastic combination of wet and dry lab expertise (Figure 1)
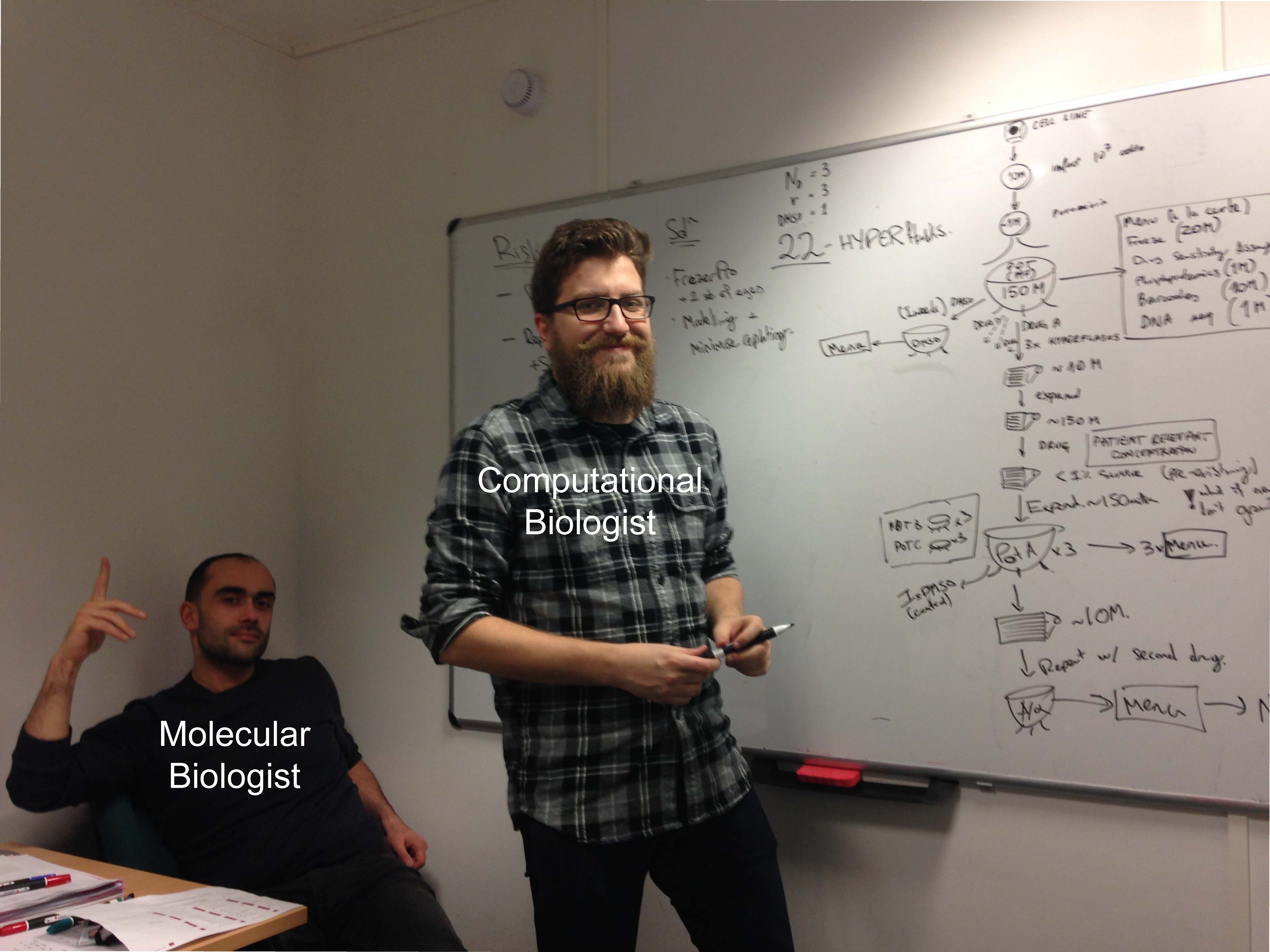
Figure 1. We put together a molecular biologist (Ahmet Acar) with a computational biologist (Daniel Nichol). A great combination of wet and dry lab expertise to do 'humid' science.
We realised that to recapitulate the temporal dynamics of tumour evolution we needed to:
- Have lots of standing variation (intra-tumour heterogeneity) using large populations. No standard plates.
- Reproduce adaptation to drugs without artificial perturbations. No usual cell passaging.
- Track cancer clones over time non-destructively.
- Use high-throughput drug screening to look for potentially rare evolutionary trade-offs.
We decided to start from a well characterise system: the lung cancer cell line HCC827. A totally uncool model.
With Hyperflasks® we cultured ~150 million cells per flask. Lentiviral barcoding allowed tracking individual clones under the selective pressure of EGFR and MEK inhibitors.
We did not passage cells. We realised that dead cells in the culture media contained readable barcodes. It turned out that these ‘floating barcodes’ are a good representation of what is alive at the bottom of the flask, allowing the monitoring of clones without ‘touching’ them.
One of the most striking results was that evolution was highly conserved between independent replicates, to the point that even the time at which clones emerged was essentially the same (see barcoded clones in Figure 2). Such predictability is a prerequisite for controllability.
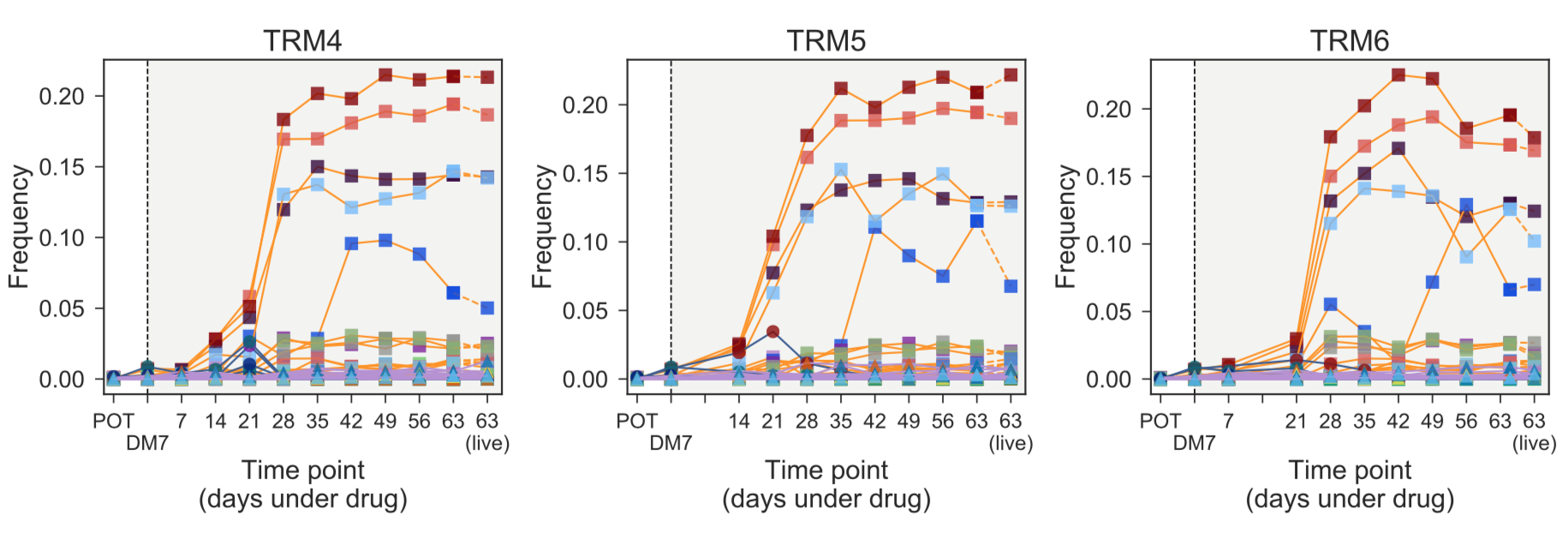
Figure 2. Temporal frequencies of the floating barcodes in three independent replicates under MEK inhibitor trametinib showed conserved evolutionary dynamics. The final samples (harvested 'live' population) largely match the last floating barcodes sample.
Resistance did come at the cost of slower growth, as well as collateral sensitivity to second-line agents, demonstrating that drugs could be used to steer the population in a controlled manner.
Our proof-of-concept study showed that evolution under the selective pressure of drugs can be highly predictable and potentially controllable. In patients, we can monitor evolution effectively with circulating tumour DNA (somehow equivalent to the floating barcodes) and then use computational and mathematical approaches to predict the next treatment step. The objective is adaptive intervention, introducing second-line drugs before a resistant tumour becomes as large and as heterogeneous as it was at the start.
There are very exciting possibilities to start validating these concepts in the clinic. Over 100 licensed anticancer drugs are used as standard of care in clinical practice and target a diverse rage of molecular targets. Using our understanding of cancer evolution, we plan to design and implement hypothesis testing clinical trials for evolutionary steering and collateral sensitivity with our colleagues at the Drug Development Unit at the ICR.
As the concept of adaptive therapy advocates (Gatenby et al. 2009), although very often in oncology we try to go for the “Rambo” approach to the problem, we may be better off going for the “Kasparov” one.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in