Cre recombinase dependent on protein X: GFP, RFP, and then?
Published in Bioengineering & Biotechnology

Recent great advances in neuroscience rely on the regulation of gene expression in specific cells. Tracing specific neuronal connections within the brain, optical imaging of genetically encoded fluorescent sensors, and optogenetics to manipulate neuronal activity in moving animals: all of them are dependent on the sophisticated regulation of gene expression in specific cells. In model animals such as mouse, Cre-loxP systems are highly prepared. Various gene-specific promoter-driven Cre mouse lines are now available for genetic access to subpopulations of cells in the brain. However, it is still difficult to create transgenic large animals including primates, and the most sophisticated genetic techniques used in mice cannot be transferred to human beings. Therefore, it is important to achieve specific gene expression control technology using viral vectors without using transgenic lines.
Nanobodies are commonly used for detection of target proteins by their very specific binding ability. They consist of a single peptide that recapitulates only the antigen binding site of the antibody molecule. The small size of nanobodies makes them ideal for integration into adeno-associated virus (AAV) vectors, which have strict genome size limitations. Recently, several methods have been developed to regulate gene expression using the target protein-specific binding properties of nanobodies. Among them, the method using split-Cre recombinase is very convenient because it can utilize existing Cre-dependent viral vectors. However, since the report of Cre recombinase dependent on GFP: Cre-DOG1, which targets GFP as its target protein, there have been no attempts to expand its target proteins.
Here, we created RFP-dependent Cre recombinases (Cre recombinase dependent on RFP: Cre-DOR) by utilizing Split-Cre reunion via specific binding of nanobodies to red fluorescent proteins (RFPs). First, eight mCherry binding proteins (MBPs) including six nanobodies and two DARPins (Designed Ankyrin Repeat Proteins) that bind specifically to mCherry were fused to the N- and C-terminal fragments of Split-Cre. Luciferase assays were then performed on 8 x 8 = 64 combinations of these 8 NCre-MBPs and 8 CCre-MBPs to find the best pair with the highest Cre recombinase activity. In experiments using GFP as a reporter of Cre-DOR recombination activity, we achieved RFP-dependent regulation of gene expression using Cre-DOR with 95% specificity in mRFP1-expressing neurons in the mouse brain. Finally, we found that local administration of Cre-DOR and reporter GFP-expressing virus vectors in Grpr-mRFP1 rats visualized the specific neural pathway from Grpr-expressing neurons in the posterodorsal medial amygdala (MePD) to the posterior bed nucleus of the stria terminalis (BSTp).
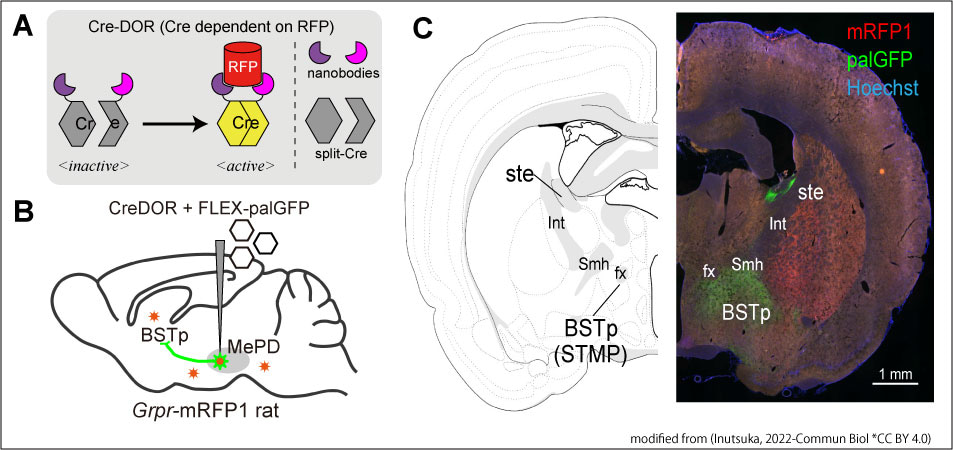 Figure 1: Schematic illustration of Cre dependent on RFP
Figure 1: Schematic illustration of Cre dependent on RFP
A, In this work, we created Cre dependent on RFP (Cre-DOR) by combining RFP-specific nanobodies and split-Cre. Specific binding of the nanobodies to RFP induces reunion of split-Cre into functional Cre recombinase. B, Local injection of virus vectors of Cre-DOR achieved specific expression of reporter palGFP in RFP-positive neurons in Grpr-mRFP1 transgenic rats. C, We revealed novel neural projections using Cre-DOR virus in RFP-expressing transgenic animals.
Now that Cre-DOG and Cre-DOR have been created, what is the next goal? We ultimately aim to create Cre-DOX (Cre recombinase dependent on protein X), a target protein X-dependent Cre recombinase for any given target protein X. Of course, many challenges remain: while GFP and RFP are almost in the same size, split-Cre reunion will be difficult if the target protein is large and the nanobody binding sites are far apart. In addition, our experiments with Cre-DOR showed that the subcellular localization of the target protein has a significant effect on Cre activity. In other words, if the target protein of interest localizes to the plasma membrane, it would be difficult to make it a target protein for Cre-DOX because Cre-DOX should be active in the nucleus. On the other hand, there is a possibility that the change in Cre activity due to such differences in subcellular localization can be utilized for detecting intracellular signals. We showed the example of glucocorticoid receptors in the Cre-DOR paper. If such a molecule that changes its subcellular localization upon signal input is used as a target protein, Cre-DOX may be able to translate intracellular signals into Cre-dependent gene expression.
Reunion of inactive protein fragments by nanobody binding is not limited to Cre recombinase. For example, it has been reported that the molecular topology of CD38 in the cell membrane can be assessed by using nanobodies that bind specifically to CD38 and split-luciferase2. Such Luc-DOX (Luciferase dependent on Protein X) will have the advantage of being able to detect membrane proteins localized at the plasma membrane and is expected to be powerful for the analysis of physiologically important molecules such as GPCRs and ion channels. A luciferase that emits near-infrared light and can be detected even in the deep brain has been reported3. Luc-DOX may enable non-invasive detection of specific target proteins in living animals including primates. Considering that machine learning using only structural information of a target protein can produce specific small-sized binding proteins, it is now very exciting to design a genetic system such as Cre-DOX or Luc-DOX.
References:
1. Cell type-specific manipulation with GFP-dependent Cre recombinase.
Tang JC, Rudolph S, Dhande OS, Abraira VE, Choi S, Lapan SW, Drew IR, Drokhlyansky E, Huberman AD, Regehr WG, Cepko CL.
Nat Neurosci. 2015 Sep;18(9):1334-41. doi: 10.1038/nn.4081. Epub 2015 Aug 10.
2. Cytosolic interaction of type III human CD38 with CIB1 modulates cellular cyclic ADP-ribose levels.
Liu J, Zhao YJ, Li WH, Hou YN, Li T, Zhao ZY, Fang C, Li SL, Lee HC.
Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8283-8288. doi: 10.1073/pnas.1703718114. Epub 2017 Jul 18.
3. Single-cell bioluminescence imaging of deep tissue in freely moving animals.
Iwano S, Sugiyama M, Hama H, Watakabe A, Hasegawa N, Kuchimaru T, Tanaka KZ, Takahashi M, Ishida Y, Hata J, Shimozono S, Namiki K, Fukano T, Kiyama M, Okano H, Kizaka-Kondoh S, McHugh TJ, Yamamori T, Hioki H, Maki S, Miyawaki A.
Science. 2018 Feb 23;359(6378):935-939. doi: 10.1126/science.aaq1067.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in