CRISPR-Cas9 off-targets guarded by short guide RNAs
Published in Bioengineering & Biotechnology

Cas9 and base editors are useful tools for making edits in DNA, programmed using guide RNAs to precisely direct them within the genome. Imagine molecular scissors, for example, cutting and correcting misprints (mutations) in our genetic code. This has been revolutionary for making laboratory models of disease and holds increasing promise as a potential medicine for cancer, sickle cell anaemia, muscular dystrophy, Huntington’s disease and many others. However, a chief concern is mis-direction of these molecular scissors to similar but incorrect sites (off-targets). AstraZeneca and others are making inroads into finding and addressing unwanted CRISPR off-targets1,2.
Our idea for reducing known off-target events was inspired by reading seminal works from Feng Zhang3, George Church4 and Jennifer Doudna5, who found that guide RNAs shorter than 16 nucleotides can direct Cas9, but do not allow nuclease activity. In other words, they form a dead Cas9 complex that shields the DNA. We thought about directing these short guide RNAs (or GUARD RNAs) to off-target sites to protect them from unwanted editing. We called this method CRISPR Guide RNA Assisted Reduction of Damage, or CRISPR GUARD.
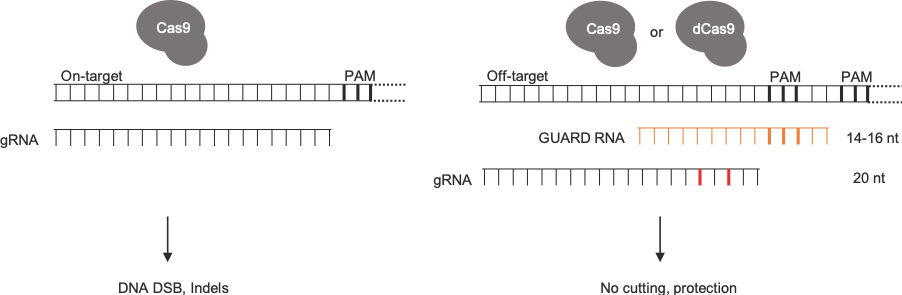
CRISPR GUARD directs inactive Cas9 complexes to shield off-target sites
After reading two fascinating studies using high-throughput systems to study Cas9 editing patterns6,7, we were compelled to adapt this approach to test the effectiveness of hundreds of GUARD RNAs. Encouragingly, this key experiment showed that most off-targets can be protected, but not unlike guide RNAs, some GUARD RNAs work better than others meaning several need to be tested. Hence, we developed an online tool to design GUARD RNAs for a given site:
https://www.sanger.ac.uk/science/tools/crisprguardfinder/crisprguardfinder/
You can find out more about CRISPR GUARD Finder here:
https://www.sanger.ac.uk/tool/crispr-guard-finder/
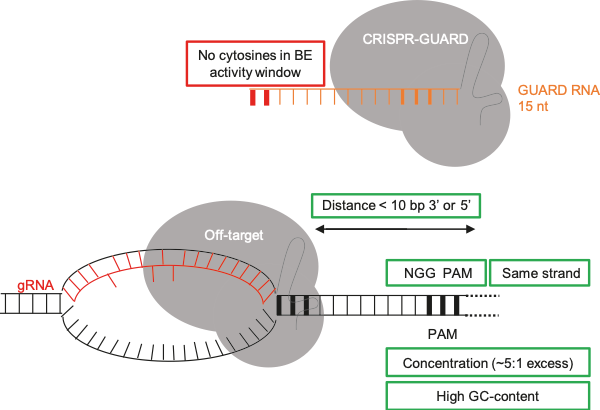
Important factors for the design of successful CRISPR GUARD RNAs
It transpired that Douglas Fowler and Dustin Maly were working on a similar idea in Seattle8 and after bumping into a co-author at a CRISPR conference in Cold Spring Harbor, New York, we decided to jointly submit to Nature Communications. It was incredible to see another group on the other side of the world independently reach similar conclusions. More recent work from our lab and elegant work from Rose, Fowler and colleagues8 show that CRISPR GUARD can be combined with high-fidelity Cas9 variants to further reduce off-target editing.
In addition, CRISPR GUARD can be used with Cas9 or base editor variants, and delivered as a ribonucleoprotein, making it easy to implement. Some obstinate or particularly detrimental off-target mutations can be eliminated completely, and multiple GUARDs together can protect multiple sites simultaneously. On-target editing is maintained in cells, probably due to limited sequence complementarity of GUARD RNAs with the on-target site, and Cas9 concentrations not becoming limiting, meaning there is enough complexed with the active gRNA. We hope CRISPR GUARD will be a useful tool for improving the specificity and safety of genome editing.
View our Nature Communications publication here:
https://doi.org/10.1038/s41467-020-17952-5
Dr. Matthew A. Coelho
Postdoctoral Fellow, AstraZeneca, Cambridge, UK
Postdoctoral Fellow, Wellcome Sanger Institute, Cambridge, UK
matthew.coelho@sanger.ac.uk
benjamin.taylor@astrazeneca.com
References
1 Akcakaya, P. et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416-419, doi:10.1038/s41586-018-0500-9 (2018).
2 Wienert, B. et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science, doi:10.1126/science.aav9023 (2019).
3 Dahlman, J. E. et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nature Biotechnology, doi:10.1038/nbt.3390 (2015).
4 Kiani, S. et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods 12, 1051-1054, doi:10.1038/nmeth.3580 (2015).
5 Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A. & Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Science Advances, doi:10.1126/sciadv.aao0027 (2017).
6 Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature, doi:10.1038/s41586-018-0686-x (2018).
7 Allen, F. et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nature Biotechnology, doi:10.1038/nbt.4317 (2019).
8 Rose, J. C. et al. Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat Commun 11, 2697, doi:10.1038/s41467-020-16542-9 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in