Deficiency of FRMD5 results in neurodevelopmental dysfunction and autistic-like behavior in mice
Published in Neuroscience and General & Internal Medicine

-
How the project start?
The origins of this research endeavor can be traced back to the intellectual discussions between Dr. Tian-Jie Lyu (specializing in neuroscience) and Dr. Ji Ma (specializing in tumor biology), both distinguished authors of this study. This journey began when they were roommates, engaging in thought-provoking conversations about their respective fields. During this time, Dr. Ma, while studying tumor biology with Frmd5 knockout (KO) mice, incidentally noticed peculiar behavioral anomalies in these mice. This casual observation sparked the inception of a research project that took root during their dormitory "fireside chats". These two doctoral students then reported the phenomenon to their respective supervisors, Professor Yun Wang and Professor Hongquan Zhang. Professor Yun Wang has been engaged in the study of the mechanism of brain diseases related to neural development for a long time, while Professor Hongquan Zhang has been focusing on research in molecular cell biology and tumor biology. After the two research groups discussed the phenomenon, they initiated a collaboration on the project. Promptly, following a preliminary behavioral assessment of the Frmd5-KO mice, they confirmed distinct abnormalities in the mice's nervous system. Consequently, the two research teams joined forces, establishing a close collaboration aimed at exploring the function of FRMD5 within the nervous system.
FRMD5, a novel member of the FERM domain-containing proteins (FDCPs) superfamily, initially discovered in 2009 as a downstream regulatory target of the p53 gene1, FRMD5 was subsequently cloned in 20122. Previous studies have hinted at the role of FRMD5 as a scaffold protein. However, the functional exploration of FRMD5 protein is still limited, primarily focusing on tumor biology2-4 and studies related to serum lipid changes in fat metabolism4-9. Additionally, a small number of reports touch upon infection-related investigations10,11.
-
ASD: a spectrum disorder with profound clinical and genetic heterogeneity
Autism Spectrum Disorder (ASD), also known as autism or autistic disorder, refers to a group of neurodevelopmental disorders that manifest in infancy and early childhood. The core symptoms include difficulties in social functioning and/or communication, restricted interests, and repetitive behaviors. ASD is considered a genetically dominated disorder influenced by environmental factors. Individuals with ASD often experience comorbidities, such as intellectual disability (ID), epilepsy, motor and sensory abnormalities, and/or attention deficit/hyperactivity disorder (ADHD)12. The substantial phenotypic heterogeneity observed aligns with the genetic heterogeneity identified over the past decade. Despite a prevalence rate of at least 1.5% in recent assessments in developed countries and the identification of numerous candidate pathogenic genes13, each specific ASD risk gene can only account for a small fraction of ASD cases (<1%). Notably, close to 50% of ASD cases lack identifiable genetic factors14,15. The phenotypic heterogeneity and genetic diversity emphasize that the term "ASD" is not a straightforward disease diagnosis; but rather a cluster of rare diseases sharing common phenotypes. Hence, comprehensive exploration of specific disrupted biological processes in distinct ASD models and understanding the resulting molecular or cellular subtypes holds tremendous potential for personalized precision treatments in the future.
-
What we found in this study?
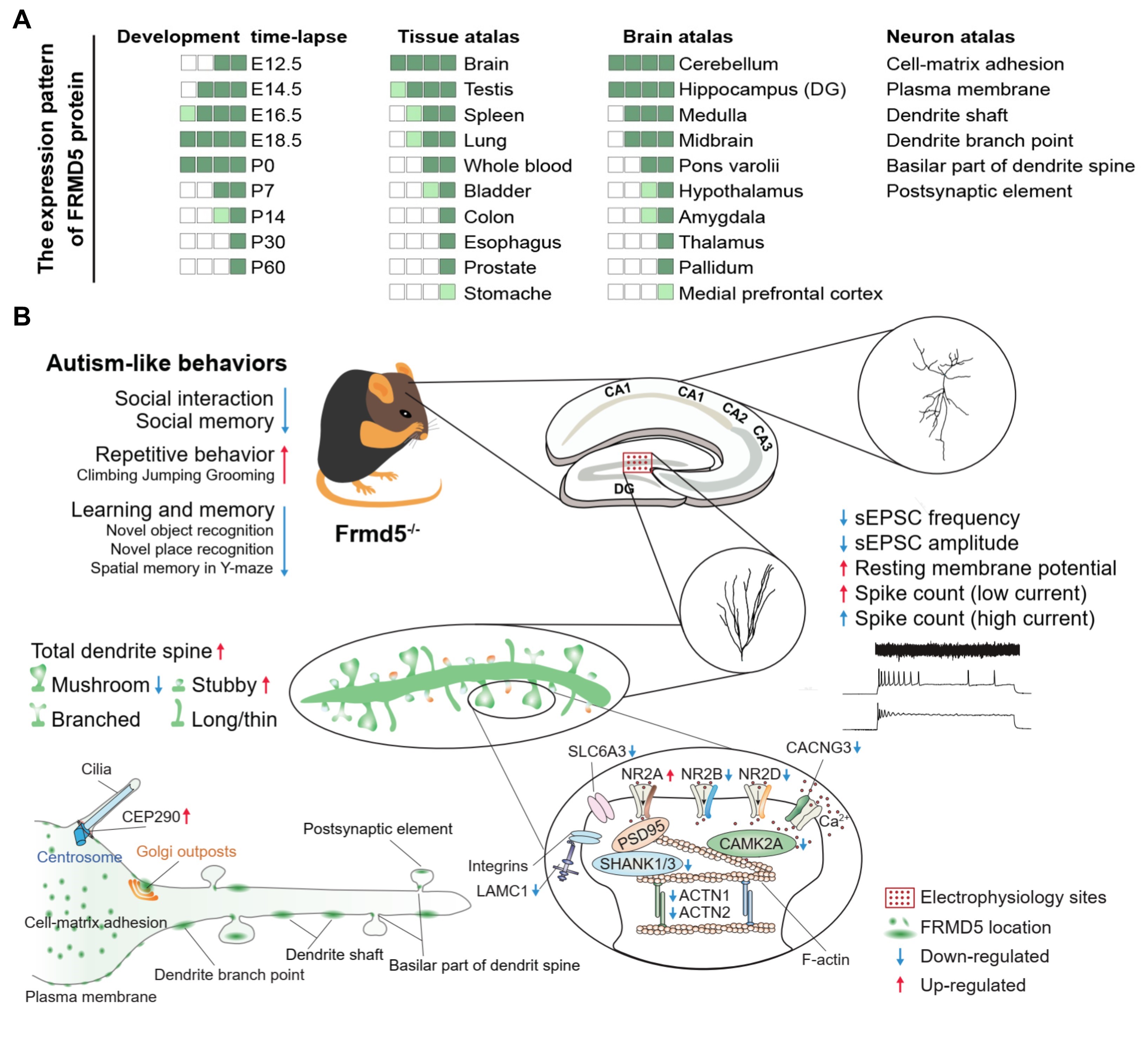
In our study, we observed high expression of FRMD5 in the mouse brain, with a peak during early embryonic development. Organ distribution analysis revealed the highest expression levels of FRMD5 in the mouse brain. Across developmental stages, FRMD5 protein exhibited a peak expression during early embryonic development in the brain. At the tissue level, FRMD5 was widely expressed in various brain regions, with particularly high expression in the hippocampus, especially the DG region. Similarly, the hypothalamus, brainstem, midbrain, and cerebellum displayed significant FRMD5 expression. Within neurons, FRMD5 in dendritic expression was prominent in dendritic shafts, branch points, and dendritic spine bases, with distribution in postsynaptic components and absence in presynaptic components. Behaviorally, Frmd5 deficiency in mice was associated with impaired learning and memory, social dysfunction, and increased repetitive stereotyped behavior. However, it did not affect ultrasonic vocalizations, motor abilities, anxiety levels, olfaction, or nociception. In terms of neurodevelopment, experiments including immunohistochemistry, sparse labeling of neurons, and Golgi staining revealed reduced dendritic branching complexity and decreased dendritic spine maturity in the hippocampal neurons of Frmd5-deficient mice. Additional experiments involving In Utero Electroporation, primary hippocampal neuron cultures, and histological analysis suggested that Frmd5 deficiency does not impact cortical neuron migration, neuron polarity, or overall brain cell tissue structure. At the neuronal functional level, electrophysiological experiments indicated increased resting membrane potential and input impedance in the dentate gyrus of the hippocampus, weakened synaptic transmission, and enhanced spontaneous excitability in Frmd5-deficient mice. At the molecular level, our TMT quantitative proteomic and Western blot experiments revealed abnormal levels of several synaptic function-related proteins in the brains of Frmd5-deficient mice, including proteins encoded by multiple ASD risk genes (CEP290, CAMK2A, SHANK1, and SHANK3), two other kinases in the CAMKs family (CAMK2G and CAMK4) associated with synaptic function, and several NMDA receptor subunits and proteins related to glutamate neurotransmitter signaling. Simultaneously, our exploration of two interactive web-based databases (DECIPHER, ClinVar) linking genotype-phenotype correlations revealed three ASD patients and four individuals with varying degrees of neurodevelopmental abnormalities, all sharing a common feature of either a single nucleotide missense mutation in FRMD5 or a substantial segmental deletion encompassing the FRMD5 gene.
-
Where to go from here?
To our knowledge, there are few studies on the functions of the FERM domain-containing protein superfamily in the nervous system. This study represents the pioneering work that comprehensively explores the involvement of the FRMD5 protein in the molecular, morphological, synaptic, and behavioral aspects of neural development and ASD pathology (Graphical abstract), shedding new light on the role of FERM domain-containing proteins in the nervous system. The generation of Frmd5 whole-body knockout mice as an ASD model holds significant implications for understanding ASD heterogeneity and provides a basis for future ASD drug development targeting FRMD5, with promising potential for clinical application. In the future, unraveling precisely which critical synaptic proteins FRMD5 anchors to key positions will be crucial. How does FRMD5 dynamically regulate its anchored proteins for efficient functioning when neurons are actively engaged in important functions? Given the distribution of FRMD5 across different neuronal structures, what are the similarities and differences in the functions of these diverse locations? Can the identification of these critical details pave the way for the design of targeted small molecules to improve clinical symptoms in patients? We are still on the exploratory journey...
References:
1 Brazdova, M. et al. Modulation of gene expression in U251 glioblastoma cells by binding of mutant p53 R273H to intronic and intergenic sequences. Nucleic Acids Res 37, 1486-1500 (2009). https://doi.org/10.1093/nar/gkn1085
2 Wang, T. et al. FERM-containing protein FRMD5 is a p120-catenin interacting protein that regulates tumor progression. FEBS Lett 586, 3044-3050 (2012). https://doi.org/10.1016/j.febslet.2012.07.019
3 Hu, J. et al. FERM domain-containing protein FRMD5 regulates cell motility via binding to integrin beta5 subunit and ROCK1. FEBS Lett 588, 4348-4356 (2014). https://doi.org/10.1016/j.febslet.2014.10.012
4 Huang, C. K. et al. Adipocytes promote malignant growth of breast tumours with monocarboxylate transporter 2 expression via beta-hydroxybutyrate. Nat Commun 8, 14706-14719 (2017). https://doi.org/10.1038/ncomms14706
5 Franceschini, N. et al. Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am J Hum Genet 91, 744-753 (2012). https://doi.org/10.1016/j.ajhg.2012.08.021
6 Desmarchelier, C. et al. The postprandial chylomicron triacylglycerol response to dietary fat in healthy male adults is significantly explained by a combination of single nucleotide polymorphisms in genes involved in triacylglycerol metabolism. J Clin Endocrinol Metab 99, E484-488 (2014). https://doi.org/10.1210/jc.2013-3962
7 Guo, T. et al. Integrative variants, haplotypes and diplotypes of the CAPN3 and FRMD5 genes and several environmental exposures associate with serum lipid variables. Sci Rep 7, 45119-45130 (2017). https://doi.org/10.1038/srep45119
8 Yeo, A. et al. Pharmacogenetic meta-analysis of baseline risk factors, pharmacodynamic, efficacy and tolerability endpoints from two large global cardiovascular outcomes trials for darapladib. PLoS One 12, e0182115 (2017). https://doi.org/10.1371/journal.pone.0182115
9 Gao, H. et al. Association of the FRMD5 rs2929282 polymorphism and serum lipid profiles in two Chinese ethnic groups. Int J Clin Exp Pathol 11, 3494-3510 (2018).
10 Masoudian, M., Derakhshandeh, A. & Ghahramani Seno, M. M. Brucella melitensis and Mycobacterium tuberculosis depict overlapping gene expression patterns induced in infected THP-1 macrophages. Iran J Vet Res 16, 368-373 (2015).
11 Wang, X. et al. Proteomic analysis of the response of Trichinella spiralis muscle larvae to exogenous nitric oxide. PLoS One 13, e0198205 (2018). https://doi.org/10.1371/journal.pone.0198205
12 Gillberg, C. & Fernell, E. Autism plus versus autism pure. J Autism Dev Disord 44, 3274-3276 (2014). https://doi.org/10.1007/s10803-014-2163-1
13 Lyall, K. et al. The changing epidemiology of autism spectrum disorders. Annual review of public health 38, 81-102 (2017). https://doi.org/10.1146/annurev-publhealth-031816-044318
14 An, J. Y. et al. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362, eaat6576 (2018). https://doi.org/10.1126/science.aat6576
15 Turner, T. N. et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am J Hum Genet 98, 58-74 (2016). https://doi.org/10.1016/j.ajhg.2015.11.023
Follow the Topic
-
Molecular Psychiatry

This journal publishes work aimed at elucidating biological mechanisms underlying psychiatric disorders and their treatment, with emphasis on studies at the interface of pre-clinical and clinical research.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcement


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in