"Defying the Odds: A Genetic Locus Confers Resistance to Bordetella Pertussis-Induced Histamine Sensitization"
Published in Healthcare & Nursing

Histamine - it's a crucial molecule that regulates various physiological processes in both humans and animals. But did you know that it's also responsible for anaphylactic shock, a severe and potentially life-threatening allergic reaction?
When the body's immune system overreacts to an allergen, it triggers the release of histamine, which can cause blood vessels to dilate, blood pressure to drop, and airways to narrow - leading to anaphylactic shock. This can result in symptoms such as difficulty breathing, rapid heartbeat, swelling of the throat or tongue, and in severe cases, loss of consciousness or even death. Understanding the genetic regulation of histamine in anaphylactic shock is critical for developing effective treatments for this serious condition.
In mice, we previously identified Histamine 1 receptor (H1R/Hrh1) as the gene regulating susceptibility to Bordetella pertussis-induced histamine sensitivity. H1R has two haplotypes in mice with the Hrh1s allele imparting hypersensitivity and the Hrh1r allele mice are resistant to histamine.
To map the entire landscape of histamine susceptibility and resistance in laboratory and wild-derived strains of mice, we genotyped the Hrh1 gene for a total of 91 strains and found strong conservation that may have been selected for functionality. Of the 91 strains, 22 carry the Hrh1r allele, whereas 69 carry the Hrh1s allele. Through serendipity, we discovered some strains of wild-derived mice such as MOLF, PWD, PWK, and others that despite carrying the Hrh1r allele turned out to be hypersensitive to histamine. This led us to hypothesize that these wild-derived strains may harbor complementing polymorphisms to override resistance to histamine. We have named this locus as an enhancer of Bordetella pertussis-mediated histamine sensitization (Bphse).
Our paper goes in depth to confirm the existence and location of Bphse using a set of genetic crosses, linkage analysis, congenic mapping, and genetic association studies. There were several key findings on this novel locus:
1) is dominant
2) requires Hrh1 to exhibit hypersensitivity to histamine
3) is linked to Hrh1 on chromosome 6
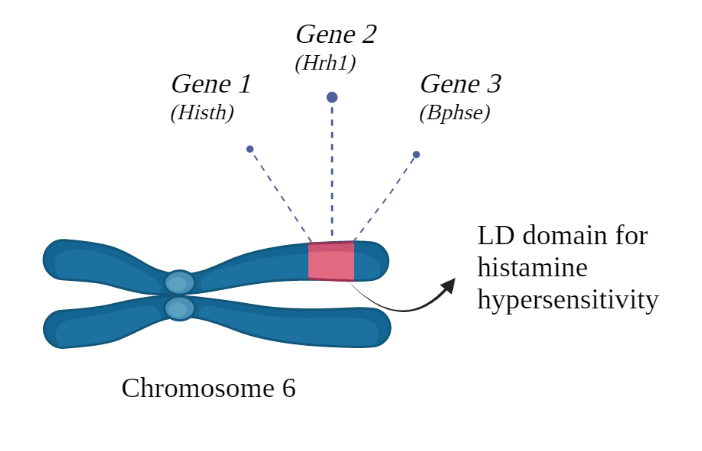
What makes our discovery even more intriguing is that we've previously identified a recessive locus (Histh) on chromosome 6 that controls age and inflammation-triggered hypersensitivity to histamine https://www.nature.com/articles/s42003-019-0647-4. With the discovery of Bphse, we've uncovered a functional linkage disequilibrium (LD) domain that encodes multiple loci controlling hypersensitivity to histamine following both Bordetella pertussis (Hrh1 and Bphse) and other factors such as age and inflammation (Histh).
Want to learn more? Check out our paper here, where we dive deep into the fascinating world of histamine sensitivity and unveil the top candidates for this dominant locus! https://www.nature.com/articles/s42003-023-04603-w#MOESM2
But that's not all - our findings have uncovered unique insights into the genetic regulation of histamine signaling, with important implications for understanding allergies and immune responses. So buckle up and get ready to be amazed as we take you on a journey through the exciting world of genetics and histamine hypersensitivity!
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in