Designing protein nanopores for capturing and degrading microplastics
Published in Bioengineering & Biotechnology

The problem
Approximately 400 million tonnes of plastic are produced globally each year, with an annual increase of approximately 4%1,2. Their manufacture is one of the contributors to climate change3. Moreover, over time, macro-plastics can undergo weathering and degradation, breaking down into smaller particles known as microplastics. These microplastics find their way into the atmosphere, oceans, wastewater and drinking water, posing substantial ecological and health challenges4-6.
Polyethylene terephthalate (PET) constitutes over 10% of the world's plastic production, yet recycling remains both scarce and inefficient7. While we can place a PET bottle in a reactor to degrade or reuse it, a pressing question arises: How can we effectively deal with nearly invisible PET microplastics? This poses a global challenge, where the aim is to develop biocatalysts not just for breaking down larger PET plastics, but also for handling sub-micro and nano-sized PET particles.
The discovery
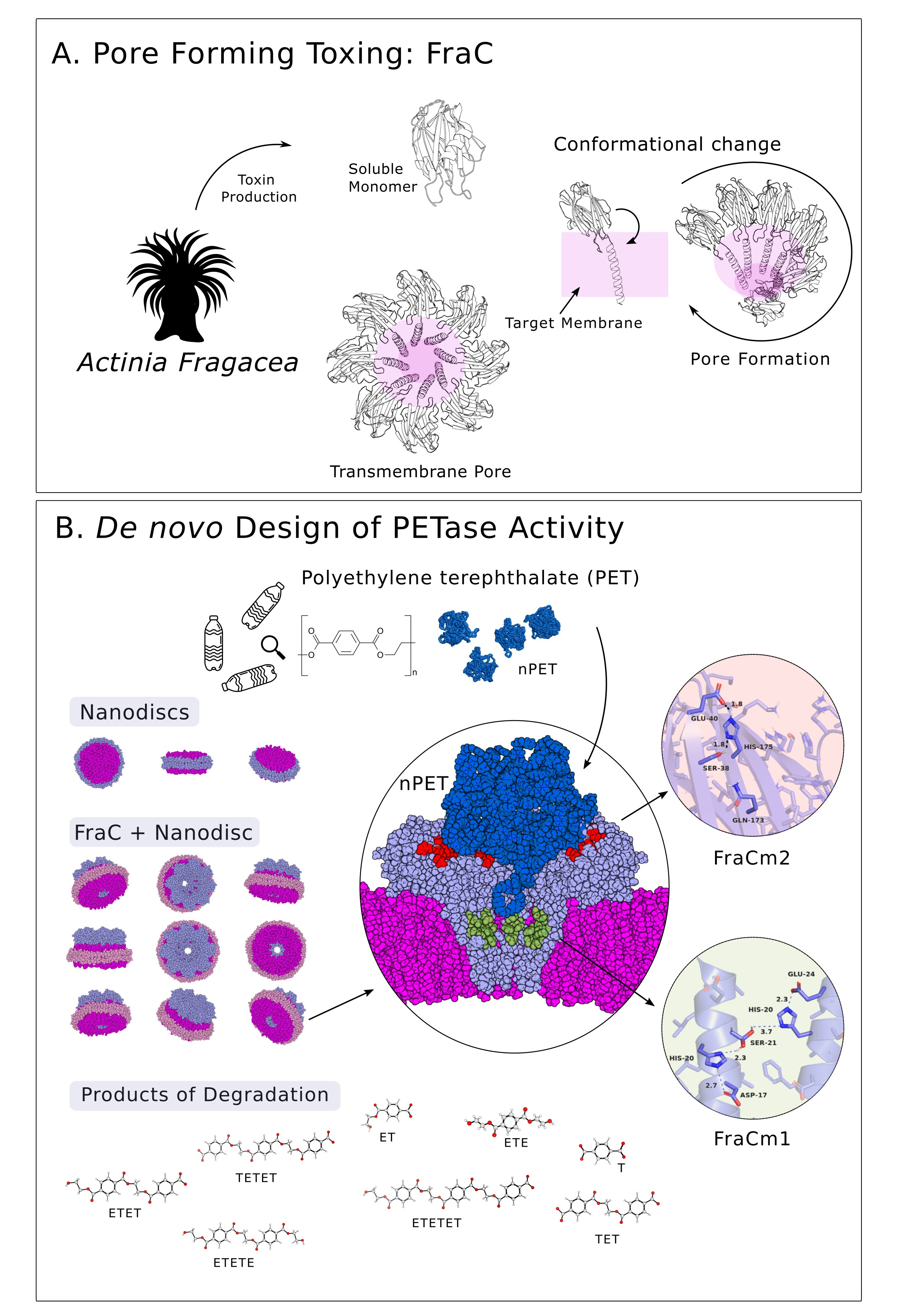
To design a protein capable of degrading microplastics, our approach is akin to enhancing a multi-purpose tool, similar to a Swiss knife, by adding new components to expand its range of functions. These complements may consist of just three amino acids that function as scissors capable of cutting small PET particles. In our study, these components have been introduced into a protein from the anemone Actinia fragacea, a protein that originally lacked this capability. A. fragacea, commonly known as the strawberry anemone, is a species of sea anemone found in shallow coastal waters. One interesting aspect of A. fragacea is its venomous capabilities, which it uses for defence and predation. Fragaceatoxin C (FraC) is a type of protein toxin found in the venom of A. fragacea. In nature, FraC comes into contact with a target cell, it can insert itself into the cell membrane and create nanopores (Figure 1A), as big as 7.0 nm high x 1.6-6.0 nm diameter, which can disrupt the integrity of the cell membrane, drilling the cell content.
The formation of nanopores by FraC is a remarkable example of how nature has evolved sophisticated mechanisms for defence. Researchers have a strong interest in examining these nanopores for their potential applications in nanotechnology, an avenue we explored in our study through the development of a FraC-based nanopore designed specifically for microplastic degradation.
Using a structure-based computational modelling approach (Protein Energy Landscape Exploration, PELE) we have introduced into FraC an artificial catalytic triad that supports ester hydrolysis. With PELE, we can anticipate the binding sites for PET particles within the FraC nanopore and strategically position the new amino acids for effective hydrolysis (see Figure 1B). The resulting structural configuration closely mirrors that of the PETase enzyme found in the bacterium Ideonella sakaiensis, discovered in a packaging recycling plant in Japan8.
Our study demonstrates that, under room temperature conditions (30ºC), the newly engineered FraC protein, when assembled into nanopores on a lipid bilayer, effectively facilitates the degradation of PET micro- and nano-plastics sourced from feedstock or plastic bottles. Alternative approaches using benchmark PETases often require temperatures above 70°C to make the plastic more flexible, contributing to elevated CO2 emissions due to the heating involved.
A nanopore design that allows for recycling and more
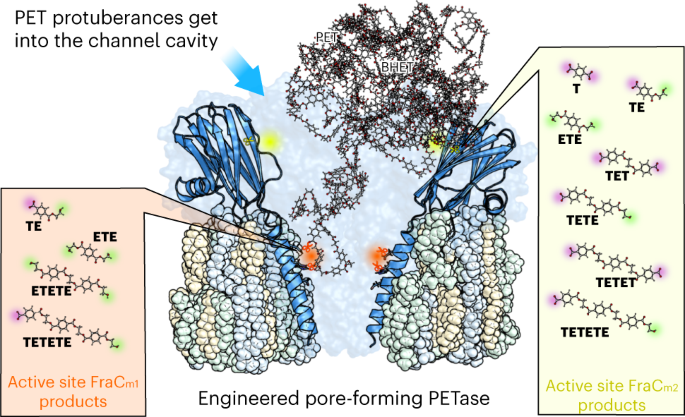
Another benefit of the novel catalytic nanopore protein, along with the opportunities offered by computational modelling techniques, lies in the design of two distinct variants based on the strategic placement of the new amino acids. The result is that each one leads to different products (Figure 1B). One variant breaks down PET particles more thoroughly, so it could be used for degradation in wastewater treatment plants. The other gives rise to the initial components needed for recycling. In this way we can purify or recycle, depending on requirements.
In addition, the protein’s pore-like structure allows water to pass through free-moving nanodiscs or be anchored to membranes similar to those used in desalination plants9. This would make it suitable for purification plants to break down PET particles that are not visible but challenging to remove, and that we may inadvertently ingest.
The versatility of the Frac protein, comparable to a 'multi-purpose tool', allows for the addition and testing of new catalytic elements and combinations. By doing so, we aim to harness the potential of protein-based nanopores found in nature and leverage supercomputing to create innovative designs that promote a plastic-free environment and advancements in the circular bio-based economy. In the first case, we can explore the application of these new designs for degrading other plastics, such as nylon. In the second case, we can consider their potential for transforming various substances, including pollutants or biomasses, such as the hydrolysis of proteins in industrial bio-waste.
Behind the paper
Based on our experience in computational analysis and hands-on exploration of natural diversity, our research in applied biotechnology is geared towards addressing PET pollution. We thought of introducing multiple hydrolysing active centers into a confined but large binding cavity; here, non-catalytic pore-forming proteins seem ideal candidates. This center consisted of an artificial catalytic triad (Ser, Asp, His) and an oxyanion hole to stabilize the charge that appears during hydrolysis; its geometry was similar to that of a benchmark PETase, thinking that by having a similar arrangement it would be possible to transform a non-catalytic nanopore into a PET-capturing and degrading one. Initial results, however, were disheartening; we were unable to identify any signature of PET film degradation, at any temperature and condition, when a first mutant was designed. Then, our research took a hopeful turn. A probing analysis of degradation tests using PET synthetic particles with sizes ranging from 53 to 154 nm (sub-micro to nano-size), revealed a substantial degradation. But how can a PET particle with a diameter of 53-154 nm fit into a nanopore with a diameter of only 1.9 (cis exit) to 6.7 nm (trans exit)? We discovered that PET particles exhibit dynamic shapes with multiple protrusions. Interestingly, it's not the particles themselves that enter the nanopores, but rather these protrusions. As the protrusions break down, the particle size decreases, reducing their ability to enter the nanopores and undergo degradation (Figure 1B). Observing this pronounced and consistent deconstruction, we explored different nanopore locations to introduce an additional active site. Depending on the placement of this artificial active center and the accessibility of the protrusions, we achieved varied degradation pathways for the nanoparticles. Thus, we found ourselves charting exciting new territory in the quest to enhance the capture and break down of sub-micro and nano-sized PET, for example, in wastewater treatment plants. M.F., A.R.-M., S.G.-L., A.M.-P., and V.G.
We acknowledge the financial support of the EU’s Horizon 2020 (GA101000327, GA101000327) and Horizon Europe (GA101060625), the MICIIN, AEI (DOI: MCIN/AEI/10.13039/501100011033), ERDF A way of making Europe and the EU NextGenerationEU/PRTR (PID2020-112758RB-I00, PDC2021-121534-I00, TED2021-130544B-I00, PID2019-106370RB-I00), UCM-Banco Santander (PR87/19-22556, PR108/20-26896) and UnaEuropa (Unano) SF2106.
References
- York A. Adapting to plastic. Rev. Microbiol. 18, 362-363 (2020).
- Rosenboom, J. G., Langer, R. & Traverso, G. Bioplastics for a circular economy. Rev. Mater. 7, 117–137 (2022).
- Cabernard, L. et al. Growing environmental footprint of plastics driven by coal combustion. Sustain. 5, 139–148 (2022).
- Allen, S. et al. Atmospheric transport and deposition of microplastics in a remote mountain catchment. Geosci. 12, 339–344 (2019).
- Schymanski, D. et al. Analysis of microplastics in drinking water and other clean water samples with micro-Raman and micro-infrared spectroscopy: minimum requirements and best practice guidelines. Bioanal. Chem. 413, 5969-5994 (2021).
- Materić, D. et al. Nanoplastics measurements in Northern and Southern polar ice. Res. 208, 112741 (2022).
- Ellis, L. D. et al. Chemical and biological catalysis for plastics recycling and upcycling. Catal. 4, 539–556 (2021).
- Yoshida, S. et al. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 (2016).
- Heiranian, M., Farimani, A. & Aluru, N. Water desalination with a single-layer MoS2 nanopore. Nat. Commun. 6, 8616 (2015).
Follow the Topic
-
Nature Catalysis

This journal brings together researchers from across all chemistry and related fields, publishing work on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, incorporating both fundamental and applied studies.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Very interesting! Thank you for sharing. I enjoyed reading and earning about this advancement while in some regions we are able to study only at the level of detection and identification of microplastics (sadly, in resource limited conditions).
learning*