A flexible and accurate AI algorithm for neurotechnologies
Published in Bioengineering & Biotechnology, Neuroscience, and Computational Sciences
Artificial Intelligence (AI) has witnessed a revolution in recent years, for example with the development of deep learning models for language, text, or images. Given its ability to capture complexity, deep learning has great potential to also benefit the modeling of brain signals. But how can we help extend the benefits of deep learning to neurotechnologies and brain-computer interfaces (BCIs) for treating neurological or even mental health conditions? This is the question we set out to answer in our new paper in Nature Biomedical Engineering.
For BCIs, we need to develop deep learning methods that address several additional challenges. First, BCIs need the ability to infer latent factors and brain states from neural population activity both causally and non-causally. For example, for offline biomarker discovery or model training, non-causal inference is important, whereas for closed-loop BCI deployment, causal inference is required. Second, real-world BCIs will be wireless and thus will need to robustly handle missing neural samples, which can happen due to wireless link interruptions. Addressing these challenges demands deep learning models that enable flexible inference, meaning the same model can support inference causally, non-causally, and with missing neural samples. Despite much progress, such flexible inference remains a challenge for deep learning models of brain data.
Motivated by this critical need, together with my PhD students Hamidreza Abbaspourazad and Eray Erturk, we developed DFINE (Fig. 1): a deep learning model of brain signals whose inference is both accurate and flexible. Further, DFINE’s inference is recursive and thus computationally efficient for real-time implementation. DFINE consists of dynamic and manifold latent factors (Fig. 1a). The dynamic latent factors describe the evolution of neural activity over a low-dimensional nonlinear manifold in tractable linear form to allow for flexible inference. The manifold latent factors characterize how this nonlinear manifold is embedded in the high-dimensional neural activity space for accuracy. We jointly train the dynamic and manifold factors as well as stochastic noise distributions by optimizing a future-step-ahead neural prediction loss. This loss was important for accurate performance and can be computed during training because of DFINE’s efficient and flexible inference (Fig. 1b).
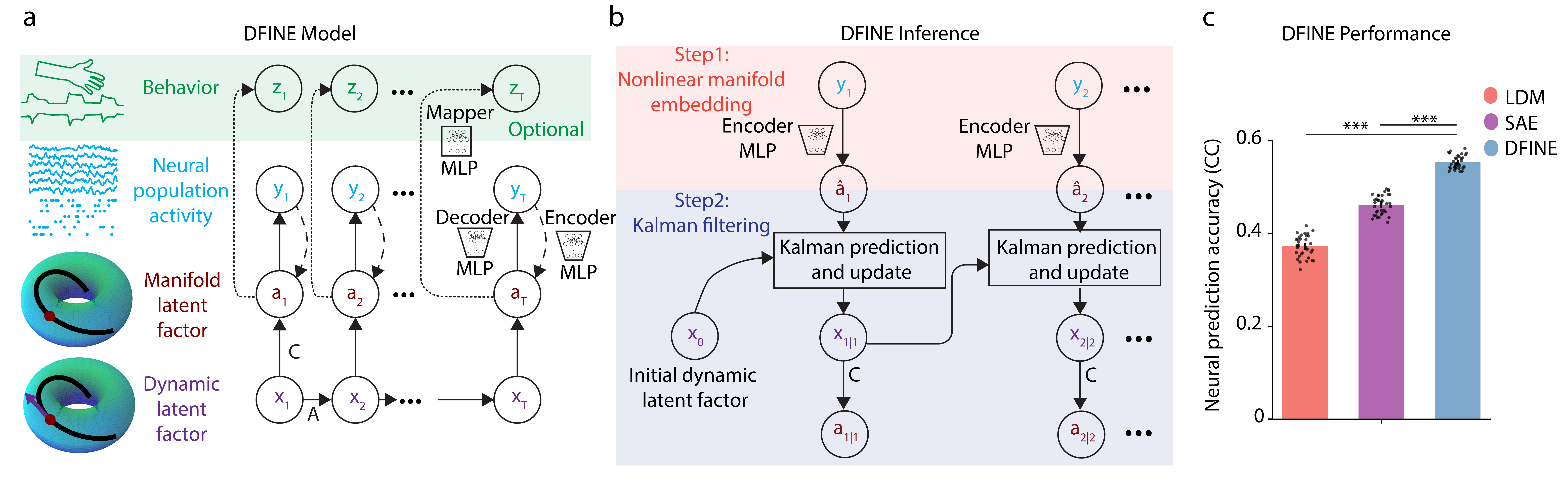
In extensive nonlinear simulations with various manifolds and with the stochastic Lorenz attractor system, we first demonstrated that the same DFINE model achieves accurate and flexible inference both causally and non-causally, and even with missing observation samples. We then showed, across multiple neural datasets with various tasks, that DFINE outperforms benchmark dynamical models of brain data, including linear dynamical models (LDM) as well as nonlinear sequential autoencoders (SAEs) (Fig. 1c). Further, we found that these improvements are larger when neural samples are missing, thus demonstrating DFINE’s robustness.
Critical to our development of DFINE were interdisciplinary insights at the interface of AI, control theory, and neuroscience. Prior work on closed-loop BCIs, including ours, largely focus on simpler computing methods including linear dynamical modeling. Linear dynamical models are particularly popular for closed-loop system design in science and engineering because of their flexible and tractable inference properties. However, linear models do not fully capture the complexity of brain signals as deep learning could, which can lead to lower accuracy. With DFINE, we achieve both flexibility and accuracy.
In the future, DFINE can help develop more accurate, robust, and effective neurotechnologies. The benefit of DFINE may go beyond traditional BCIs for decoding and extend to BCIs for closed-loop regulation of abnormal brain activity patterns1. Indeed, one focus in my lab is to develop next-generation BCIs for treating mental health conditions, such as major depression, by working at the interface of AI and control theory as I describe in a Perspective in Nature Neuroscience1. Such a BCI would decode mental states such as mood symptoms, as we demonstrated in a paper in Nature Biotechnology2. It could then use this decoded state as feedback to regulate abnormal brain activity patterns with input, such as deep brain stimulation therapy, whose effect we modeled in another paper in Nature Biomedical Engineering3. One question for future investigation is whether and how DFINE can model the effect of inputs and enable the development of closed-loop control methods for precise regulation of brain activity. Another question for future work is how to extend DFINE to different modalities of neural data, whether electrical, optical, or acoustic, or even to multiple modalities at the same time.
We are very excited about what the future holds for interdisciplinary work at the nexus between AI, engineering, and neuroscience, and how such work may lead to neurotechnologies that could transform our understanding and treatments of brain disorders.
References
- Shanechi, M. M. Brain-machine interfaces from motor to mood. Nature Neuroscience 22, 1554–1564 (2019).
- Sani, O. G. et al. Mood variations decoded from multi-site intracranial human brain activity. Nature Biotechnology 36, 954–961 (2018).
- Yang, Y. et al. Modelling and prediction of the dynamic responses of large-scale brain networks during direct electrical stimulation. Nature Biomedical Engineering 5, 324–345 (2021).
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcement

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in