Discovering the hidden: Advancing antibiotic resistance detection with real-time genomics
Published in Microbiology
The threat of antibiotic resistance
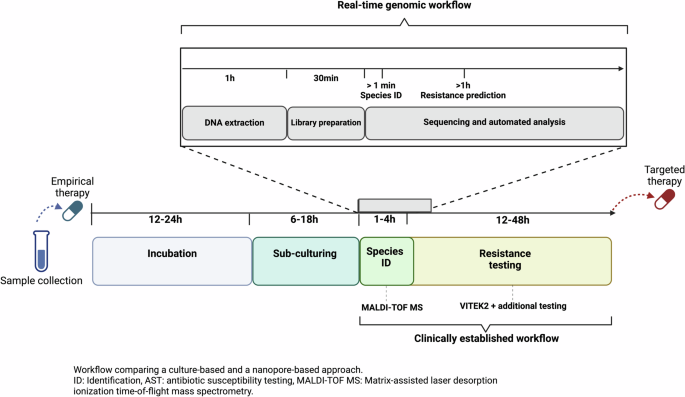
The battle against antibiotic resistance poses one of the most significant challenges in modern healthcare, threatening to render many of our most potent medicines ineffective. This global health crisis not only compromises patient outcomes but also burdens healthcare systems with increased costs and longer hospital stays. Conventional diagnostic methods to identify a pathogen’s resistance profile are primarily rooted in culturing and phenotypic testing, which often fall short in their ability to rapidly and accurately detect antibiotic resistance, particularly when it comes to rare or low-abundance resistance. The inherent delays in growing cultures and the subsequent testing for phenotypic antibiotic susceptibility can lead to critical delays in administering effective treatment, significantly impacting recovery and survival rates.
Real-time genomics in clinical diagnostics
Our recent study, peer-reviewed via Nature Communications, highlights significant advancements in the field of real-time genomics through nanopore sequencing. We were able to show that nanopore sequencing has the potential to surpass traditional diagnostic tools, providing a faster, more in-depth analysis of pathogen genomes and resistance profiles.
The focus of our study was a clinical case involving a severe infection caused by multi-drug resistant Klebsiella pneumoniae, a pathogen commonly encountered in hospital settings and known for its rapid acquisition and transmission of resistance genes. Initially, the patient was treated with Ceftazidime-Avibactam, an antibiotic combination aimed at combating such resistant infections. This choice of treatment was based on the results of phenotypic antibiotic susceptibility testing, which indicated that the pathogen was susceptible to this drug regimen.
However, the patient's condition did not improve, and subsequent blood cultures taken after the initial treatment grew K. pneumoniae that now exhibited resistance to Ceftazidime-Avibactam, indicating a significant shift in the pathogen's antibiotic resistance profile. Crucially, conventional diagnostics could not identify any specific genetic mechanisms responsible for this newfound resistance, highlighting a significant limitation of these traditional methods. It was only through the deployment of real-time genomics through long-read nanopore sequencing that the team could uncover a complex array of resistance mechanisms, including a previously undetected novel gene variant explaining the emergence of Ceftazidime-Avibactam resistance under antibiotic exposure. Importantly, this novel plasmid-encoded resistance gene was already detected in low abundance in the pathogen’s genetic material prior to Ceftazidime-Avibactam exposure, highlighting how the resistance-encoding plasmids might have risen in frequency due to selection pressure under Ceftazidime-Avibactam exposure, subsequently resulting in phenotypic resistance. The detection of the low-abundance plasmid and resistance gene by real-time genomics before Ceftazidime-Avibactam exposure could have elucidated the pathogen’s adaptation potential under selection pressure, which emphasizes the necessity for continuous and real-time genomics-based surveillance of antibiotic resistance in the clinic.
General clinical and public health implications
The integration of real-time genomics into clinical practice marks a shift in how we could manage infectious diseases in the future, both at the individual and community levels. By enabling rapid, on-site sequencing of pathogens, this technology empowers healthcare providers to make informed decisions quickly, tailoring treatment strategies to the specific genetic profile of the infecting organism. For individual case management, this immediate and in-depth insight into resistance profiles allows clinicians to prescribe the most effective antibiotics, reducing the trial-and-error approach that can delay or even prevent recovery. By ensuring that treatment is both specific and appropriate, this technology not only enhances patient outcomes but also minimizes the unnecessary exposure to broad-spectrum antibiotics, thus reducing the risk of further resistance emergence.
Beyond infection case management, real-time genomics-based antibiotic resistance detection can be leveraged for effective infection control and containment strategies. The short turnover time coupled with high resolution of resistance mechanism detection allow for targeted and immediate implementation of containment measures,potentially significantly reducing the risk of resistance transmission. This is especially crucial in settings with a highburden of antibiotic resistance, where rapid intervention can change the course of disease outbreaks.
Impact and Outlook
Our study demonstrates the capabilities of real-time genomics in detecting hidden antibiotic resistances with potentially significant impacts on our approach to diagnosing and treating infectious diseases. This technology not only allows us to gain a deeper understanding of how pathogens adapt and develop resistance but also provides a template for how continuous, real-time surveillance of antibiotic resistance can be implemented. Embracing these advanced technologies is crucial for enhancing healthcare outcomes and is pivotal in our ongoing battle against antibiotic-resistant pathogens. As we move forward, the integration of real-time genomic diagnostics into routine clinical practice offers promising solutions that could reshape our global health landscape.
As real-time genomic analysis becomes more accessible and cost-effective, it is set to influence how we detect and monitor antibiotic resistance globally: The decreasing cost and increasing accuracy of real-time genomics have broadened their application, facilitating the widespread adoption of genomic tools beyond high-resource hospital settings. The continuous evolution of this technology promises to enhance our ability to effectively combat infectious diseases, aligning with global efforts to curb the spread of antibiotic resistance.
For those interested in the detailed findings and methodologies of this study, the full paper is available here.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in