Discovery of new resistance pathway in Lung Cancer through comprehensive Phospho-proteomic analysis
Published in Cancer

Purpose of the project
High prevalence of FGFR1 amplification in lung cancer made it an interesting and promising target, especially in those lung cancer types, which lack molecular targets. Consequently, various inhibitors have been synthesized to target FGFR1 gene amplification specifically (e.g., AZD4547 and BGJ398).1 Specific FGFR1 inhibitors have been tested in clinical trials phases I and II. Clinical trials have shown promising results where the inhibitors could achieve disease-control rate of 26–39%. Moreover, some patients have benefited from the treatment for more than 14 months, which is quite promising keeping in mind that those patients were at the last stage of lung cancer with bad prognosis and short predicted survival. Nonetheless, a sizable group of patients have shown resistance to FGFR1 inhibition.2,3
Different compensation mechanisms have probably allowed lung cancer cells with FGFR1 gene amplification to escape FGFR1 inhibition. These resistance mechanisms were either originally activated in the targeted cells (intrinsic resistance) or induced throughout the course of the treatment (induced resistance). Therefore, the success of FGFR1 inhibition as a targeted therapy in lung cancer depends on our ability to characterize, understand and overcome these resistance mechanisms in order to achieve sustainable response to FGFR1 inhibition.
In our current research project, we aimed to characterize the different mechanisms of resistance to FGFR1 inhibition in squamous cell lung cancer. We intended to achieve our goal through performing a wide-scale phosphoproteomic screening using high-resolution mass spectrometry combined with RNA and DNA sequencing.
What is new in our approach?
To the best of our knowledge, the current study proposes the first wide-scale phosphoproteomic screening of three types of resistance against FGFR1 inhibition in lung cancer cells, i.e., (i) intrinsic, (ii) pharmacologically induced and (iii) mutationally induced resistance compared to sensitive cells. Additionally, results of the screening were correlated and validated through staining human tissue samples of 175 lung cancer patients.
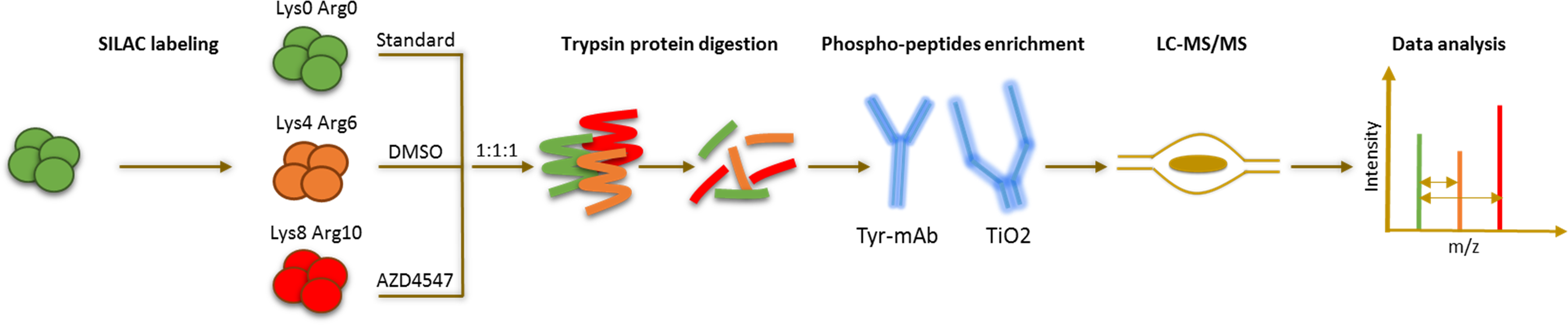
What did we discover?
We were able to identify a new signaling axis that led to FGFR1 inhibition resistance in lung cancer cells. The resistance pathway consisted mainly of the three proteins CD44, PAK1 and AKT. Combination inhibition of AKT and FGFR1, CD44 and FGFR1 or PAK1 and FGFR1 could reverse the resistance and re-sensitize intrinsically resistant and induced-resistant lung-cancer cells synergistically to FGFR1 inhibition. Additionally, squamous cell lung cancer patient tissue samples could validate the correlation between strong CD44 expression and AKT activation in SQCLC patients. Finally, our phosphoproteomic screening datasets alongside RNA and DNA sequencing could provide a valuable library for studying resistance-associated phosphorylation patterns in lung cancer cells.
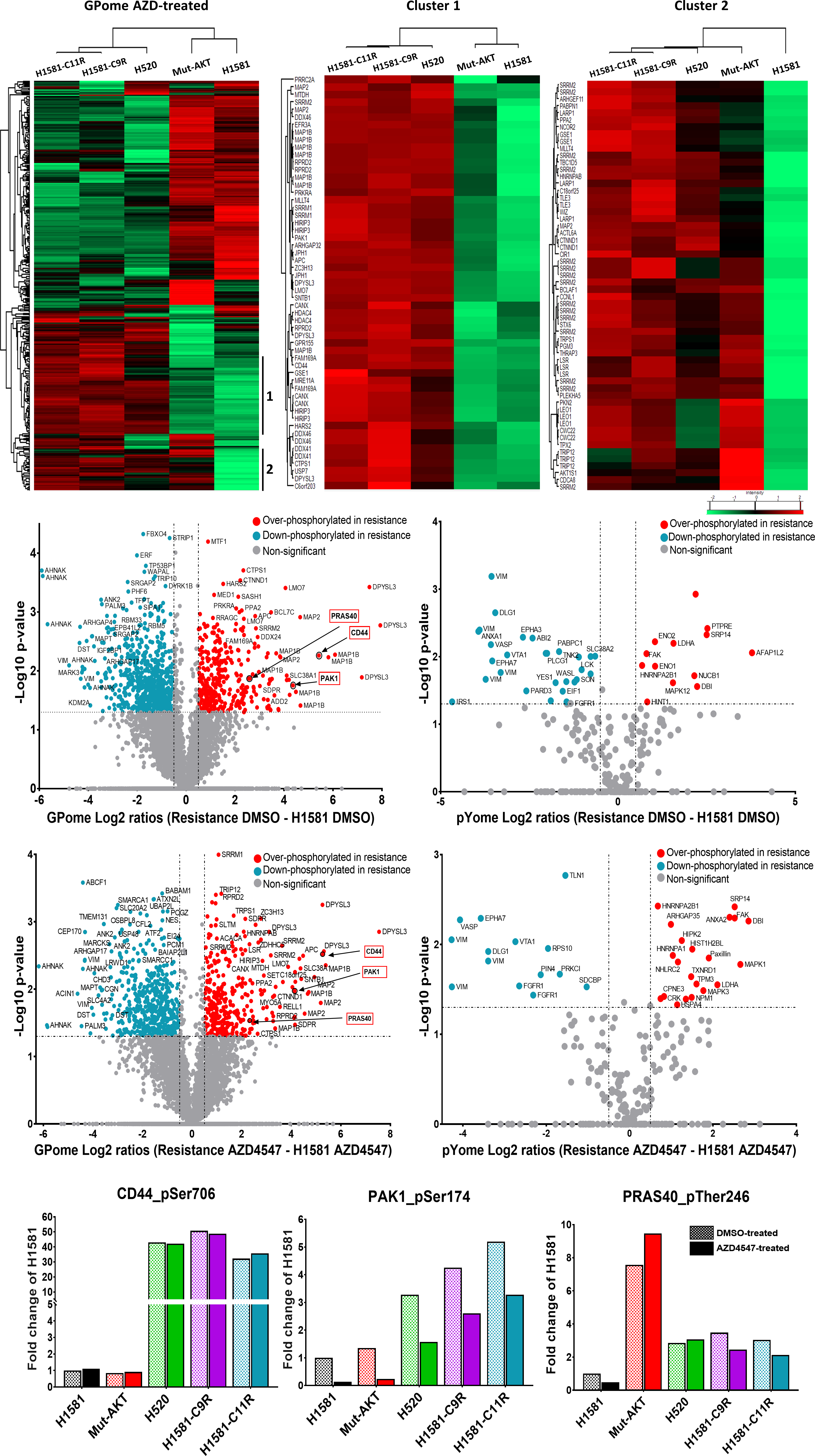
Comprehensive phospho-LC-MS/MS analysis comparing lung-cancer cells that are sensitive to FGFR1 inhibition with those that are resistant
Translational relevance of our discovery
On the one hand, the current project provides a wide-scale phosphoproteomic library of protein regulations that stand behind intrinsic and induced resistance against tyrosine kinase inhibition in cancer targeted therapy. The library introduces protein phosphorylation patterns in sensitive and resistant cell lines under control and treatment conditions, which could be used as a reference for FGFR1 inhibition specifically and tyrosine kinase inhibitors in lung cancer generally. On the other side, the project reveals a mutual resistance pathway between different types of resistance developed against FGFR1 tyrosine kinase inhibitors. The resistance pathway is composed mainly of CD44 overexpression / activation that led to PAK1 expression / activation and AKT activation. Examining levels of CD44 and/or AKT activation might predict the response of lung cancer patients to FGFR1 targeted treatment. Moreover, combinational therapy between CD44/PAK1/AKT and FGFR1 can significantly and synergistically reverse this behavior and sensitize resistant cells to the tyrosine kinase inhibition.
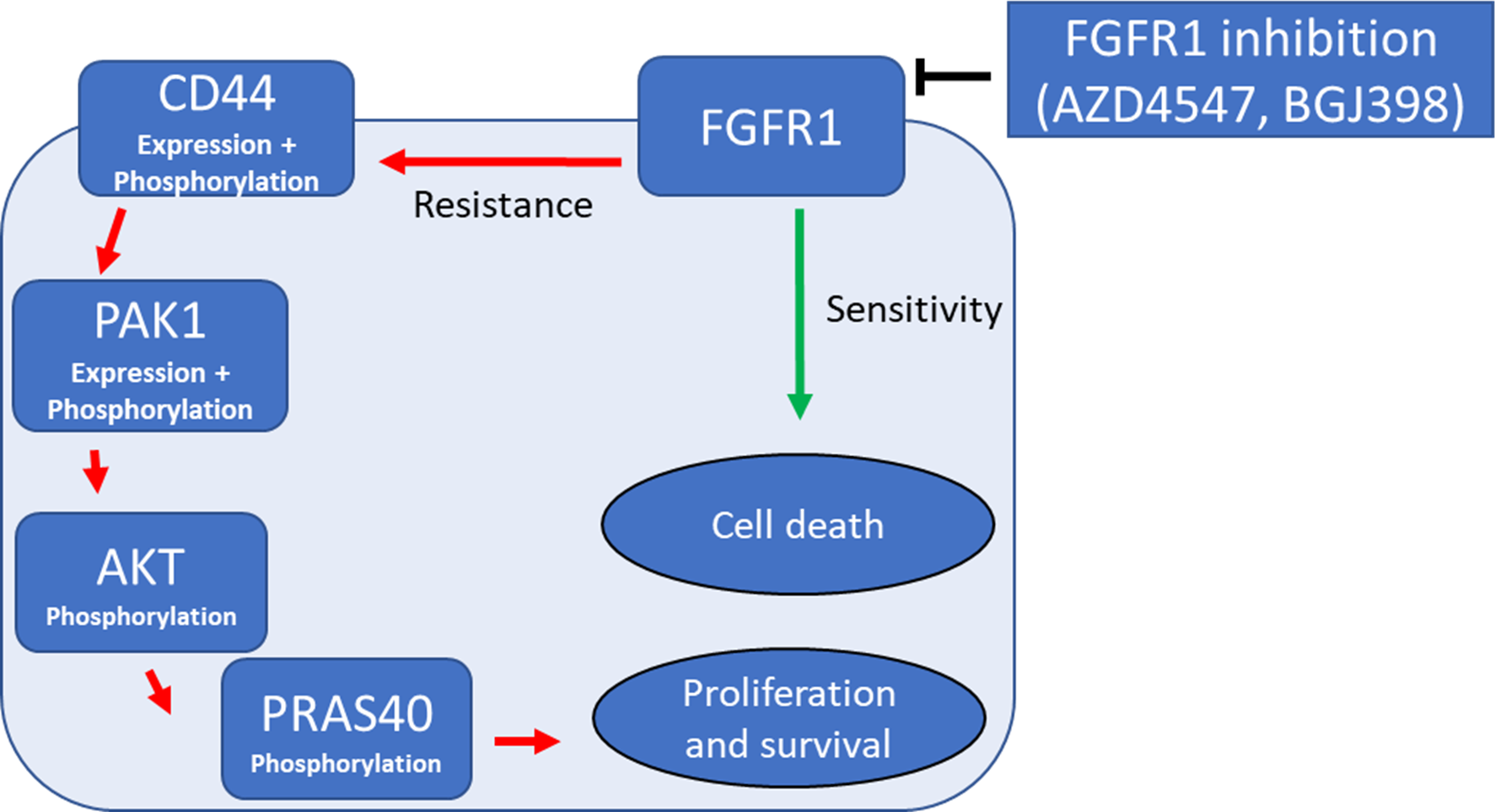
Proposed resistance axis to FGFR1 inhibition in lung cancer cells
References
- Katoh, M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat Rev Clin Oncol 16, 105-122, doi:10.1038/s41571-018-0115-y (2019).
- Aggarwal, C. et al. SWOG S1400D (NCT02965378), a Phase II Study of the Fibroblast Growth Factor Receptor Inhibitor AZD4547 in Previously Treated Patients With Fibroblast Growth Factor Pathway-Activated Stage IV Squamous Cell Lung Cancer (Lung-MAP Substudy). J Thorac Oncol 14, 1847-1852, doi:10.1016/j.jtho.2019.05.041 (2019).
- Nogova, L. et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol 35, 157-165, doi:10.1200/JCO.2016.67.2048 (2017).
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
AI Approaches in Drug Design
Publishing Model: Open Access
Deadline: Mar 31, 2026
Genomic Instability
Publishing Model: Open Access
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in