Diversity in taste perception in vertebrates driven by the complex evolution of taste receptor genes
Published in Ecology & Evolution, Neuroscience, and Cell & Molecular Biology

Humans perceive five basic tastes: umami (savory), sweet, bitter, sour, and salty. Taste buds on the tongue are responsible for detecting these tastes. Each taste cell within a taste bud expresses a specific taste receptor that recognizes distinct chemical compounds.
In 1908, Japanese chemist Dr. Kikunae Ikeda introduced the term “umami” to describe the savory taste sensation, in addition to the other four known tastes (Ikeda, 1909; translated in 2002). The word “umami” is derived from the Japanese words “Umai” meaning “delicious,” and “-mi” meaning “taste”. Ikeda identified monosodium glutamate as the key chemical compound responsible for eliciting the umami taste. Nearly a century following Ikeda’s groundbreaking work, the taste 1 receptor (T1R) family, a group of the G protein-coupled receptor superfamily, was elucidated as the primary mediator of umami and sweet taste perception (Nelson et al. 2001; Nelson et al. 2002). The human T1R family comprises three members: T1R1, T1R2, and T1R3, encoded by the TAS1R1, TAS1R2, and TAS1R3 genes, respectively. The T1R1+T1R3 heterodimer is responsible for umami taste perception, while the T1R2+T1R3 heterodimer mediates sweet taste detection. This pattern is conserved in zebrafish and medaka, where T1R1+T1R3 and T1R2+T1R3 form heterodimers to bind ligands. Therefore, it has been widely accepted that the common ancestor of vertebrates possessed these three TAS1R genes, which have been vertically transmitted to extant species.

Kikunae Ikeda (1864–1936), the discoverer of umami. Courtesy of the Department of Chemistry of the University of Tokyo.
I, too, subscribed to this conventional wisdom without question until several years ago. My initial interests lay in the evolutionary roots of varied taste perceptions among mammals, prompting me to delve into T1R analysis in mammals and several vertebrates. During the analysis, coelacanth, bichir, and elephant fish were incorporated into the dataset as outgroups, and I observed a few TAS1R sequences that were unexpectedly positioned in the phylogenetic tree. While this seemingly insignificant observation did not impact the project's progress and could have been overlooked, I sensed that it held the potential to unlock an intriguing aspect of the evolutionary history of vertebrate taste perception. This hunch propelled me to embark on a thorough investigation.
We meticulously extracted TAS1R gene sequences from the genomic and transcriptomic data of a taxonomically diverse array of vertebrates and conducted a comprehensive phylogenetic analysis. The results were compelling enough to challenge our conventional understanding, revealing that TAS1R genes in vertebrates were classified into 11 distinct clades. The newly identified TAS1R genes are expressed in the taste buds of bichir, suggesting their role as taste receptors. Evolutionary analysis of TAS1R genes revealed that following their emergence in the common ancestor of jawed vertebrates, gene duplication events led to an expansion to five distinct genes. Following the divergence of cartilaginous fish, additional gene duplication events in the common ancestor of bony vertebrates gave rise to nine distinct TAS1R genes. This suggests that our ancestors likely possessed the ability to perceive a diverse range of detectable flavors. Subsequently, TAS1R genes underwent independent loss in each lineage, resulting in the retention of only three genes in humans.
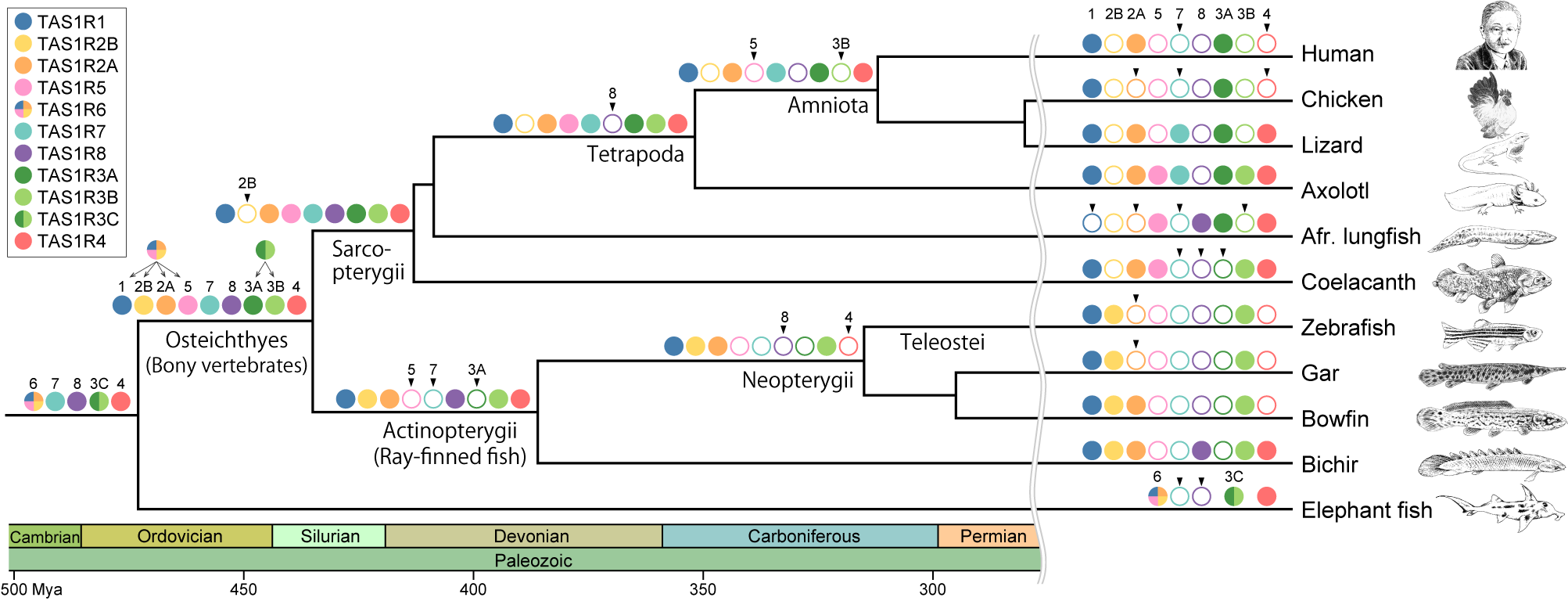
Evolution of the TAS1R family during vertebrate evolution.
The color key indicates the names of the various TAS1R members. Filled, colored circles on the branches indicate the presence of the TAS1R members, whereas open circles indicate their absence. Arrowheads above open circles indicate that the TAS1R member was lost at the branch.
Moreover, our cell-based functional assay in cultured cells successfully identified ligands for several T1Rs in bichir and elephant fish. These newly discovered T1Rs in both species, including bichir T1R8+T1R4 and elephant fish T1R6-2+T1R4, demonstrated the ability to detect branched-chain amino acids like valine and leucine, whereas mammalian and teleost T1Rs exhibit minimal or no response to these ligands. Notably, bichir T1Rs was shown to respond to amino acids essential for fish nutrition, suggesting that this fish may perceive essential amino acids as detectable. Unexpectedly, the T1Rs in elephant fish also showed similar functional characteristics, implying that these characteristics may reflect ancestral functions inherited from the common ancestor of jawed vertebrates.

This species possesses the newly discovered T1Rs, such as T1R4 and T1R8. Our cell-based functional analysis revealed that the pairs T1R1+T1R3B, T1R2B+T1R3B, and T1R8+T1R4 form heterodimers and respond to nutritionally essential amino acids.
Reexamining the evolution of T1Rs, a topic previously considered settled, has yielded a wealth of novel insights. This experience underscores the crucial lesson that non-model organisms harbor valuable evolutionary clues within their genomes. Additionally, this study has unveiled a plethora of new associated enigmas. One perplexing question surrounding TAS1R gene loss is how our ancestral species managed to survive despite losing a significant number of their T1R taste receptors. While a definitive answer to this question remains elusive, I am confident that future evolutionary studies and functional analyses of T1Rs in diverse vertebrate species will shed light on this intriguing mystery.
References
Ikeda, K. New seasonings. Chem Senses 27, 847-9 (2002).
Nelson, G., Hoon, M.A., Chandrashekar, J., Zhang, Y., Ryba, N.J. & Zuker, C.S. Mammalian sweet taste receptors. Cell 106, 381-390 (2001).
Nelson, G., Chandrashekar, J., Hoon, M.A., Feng, L., Zhao, G., Ryba, N.J. & Zuker, C.S. An amino-acid taste receptor. Nature 416, 199-202 (2002).
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in