DOLPHIN Reveals the Hidden Landscape of Exon-Level Variation in Single Cells
Published in Bioengineering & Biotechnology, Computational Sciences, and Genetics & Genomics
The Hidden Signals in scRNA-seq
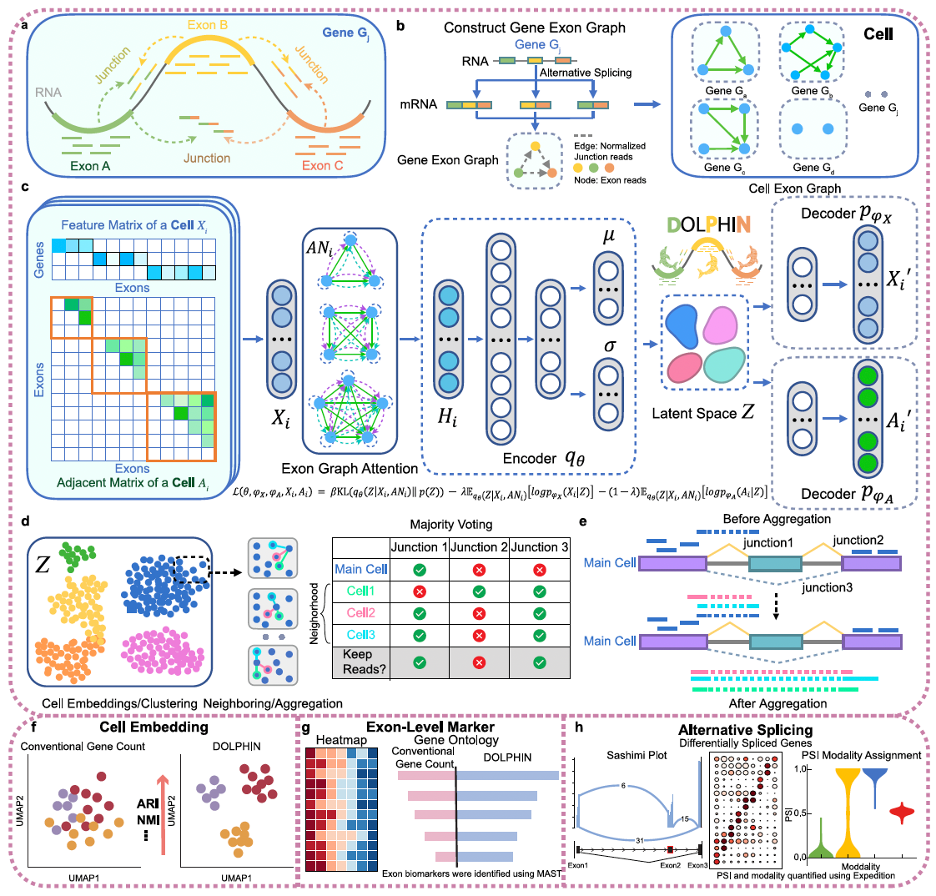
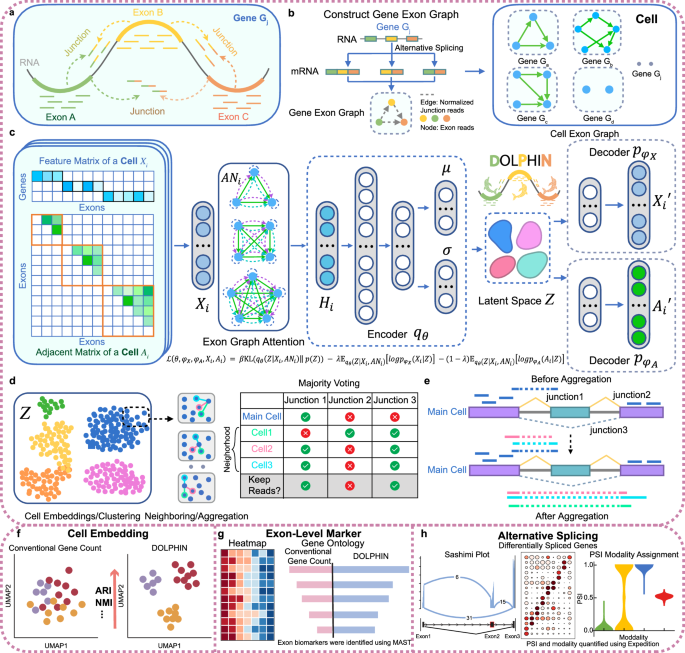
Most single-cell RNA-seq analyses treat each gene as a single count, capturing only the tip of the iceberg and missing the wealth of information hidden beneath the surface. This narrow view overlooks the fine-grained patterns that define a cell’s true identity. In contrast, DOLPHIN works at higher resolution by modeling how transcripts are assembled in each cell and capturing the subtle connections between exons, drawing on information from both exon reads and junction reads. By integrating these patterns with splicing signals, DOLPHIN creates richer cellular profiles that reveal differences others often miss, from fine splicing variations to hidden biological signatures. These enhanced profiles improve downstream tasks such as cell clustering and biomarker discovery and remain robust even for sparse datasets such as 10X Genomics. The study shows how DOLPHIN uncovers biological signals with clear potential for clinical impact.
Unlocking Hidden Signals with DOLPHIN
Building on these innovations, DOLPHIN delivers capabilities that go beyond the reach of conventional single-cell RNA-seq analysis, turning its higher-resolution view into concrete biological and clinical insights. First, DOLPHIN produces richer and more accurate cell representations, enabling sharper clustering and more precise cell type annotation than standard gene-level tools, even with sparse data from platforms such as 10X Genomics. Second, it detects exon-level differential expression signals that conventional analyses completely miss and reveal novel biological insights for better understanding of the disease. For example, in pancreatic ductal adenocarcinoma (PDAC), DOLPHIN uncovered more than 800 exon-level markers invisible to gene-level methods, including well-known cancer drivers such as SMAD4, ATM, and ERCC1. These markers not only align with established cancer biology but also separate high-risk from low-risk patients in The Cancer Genome Atlas with strong statistical significance, revealing clinical signals hidden from standard approaches. Third, DOLPHIN excels at alternative splicing analysis, a capability that conventional gene-level methods simply cannot offer. By aggregating splicing information from similar cells, DOLPHIN makes it possible to detect alternative splicing events even in sparse single-cell data, and it consistently outperforms other splicing-aware tools in both sensitivity and accuracy. For example, it detected a CD45 splicing event that distinguishes naïve from memory T cells — a fine-scale pattern missed by both gene-level analyses and existing AS tools. These advances open new possibilities for exploring cell diversity, discovering clinically relevant biomarkers, and uncovering disease-related transcriptomic regulation.
Turning Hidden Signals into Discovery
Beyond specific use cases, DOLPHIN has demonstrated unmatched robustness across diverse single-cell datasets, sequencing platforms, and tissue types. It consistently outperforms both traditional gene-level tools and recent splicing-aware methods, delivering richer cell representations and clearer separation of cell types. More importantly, by integrating exon reads and junction reads, DOLPHIN captures a much richer cellular information than traditional gene-level single-cell analyses. This added detail produces higher-resolution cell representations that preserve subtle exon-level differences between cells and uncover fine alternative splicing patterns, which are otherwise masked at the gene level. Virtual cells built on these enhanced representations can more accurately reflect the molecular state of each cell. With such high fidelity cellular models enabled by DOLPHIN, researchers can more effectively simulate disease progression at the cellular level, explore how transcriptomic changes drive pathology, and evaluate potential interventions in silico. In turn, this can deepen our understanding of disease mechanisms, improve diagnostic precision, and guide the development of more targeted and effective treatments.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in