Enabling FAIR Data in Language Cognition: Introducing HED LANG.
Published in Neuroscience and Research Data
FAIR (Findable, Accessible, Interoperable, Reusable) data transforms single-use datasets into long-term resources, enabling more transparent and reproducible science. By increasing the impact and reusability of data, FAIR principles empower researchers to compare their work with existing studies and adopt innovative methods. Recognizing these benefits, journals and funding agencies are now requiring data to be shared in open and FAIR formats.
Metadata plays a key role in making data FAIR. According to the FAIR principles, metadata should be richly described, stored in accordance with accepted formats, and should make use of standardized vocabularies. However, adhering to these principles is not always achievable. Research data is often acquired in highly specific contexts, and suitable standards to annotate such data are not always available. This creates significant barriers to making data FAIR, particularly in complex fields like cognitive neuroscience.
Cognitive science draws from diverse domains — including biology and linguistics — to understand human perception and cognition. The study of language cognition bridges the gap between linguistic theory and cognitive processes. Researchers in this field use variations in linguistic materials to elicit changes in cognition, measured through behavioral outcomes (e.g., reaction time) or neuroimaging data. For data collected in this domain to be findable, interoperable, and reusable, the linguistic properties of the presented material must be described in a standardized, machine-readable format. Without this standardization, researchers face significant hurdles in sharing, comparing, and reusing data across studies and domains.
In our recently published article, HED LANG – A Hierarchical Event Descriptors Library Extension for Annotation of Language Cognition Experiments, we introduce a new standard for annotating cognitive language experiments. HED LANG extends the existing Hierarchical Event Descriptors (HED) Schema, enabling researchers to annotate linguistic events and stimuli in a standardized, machine-actionable way. This extension addresses the unique challenges of making language cognition data FAIR.
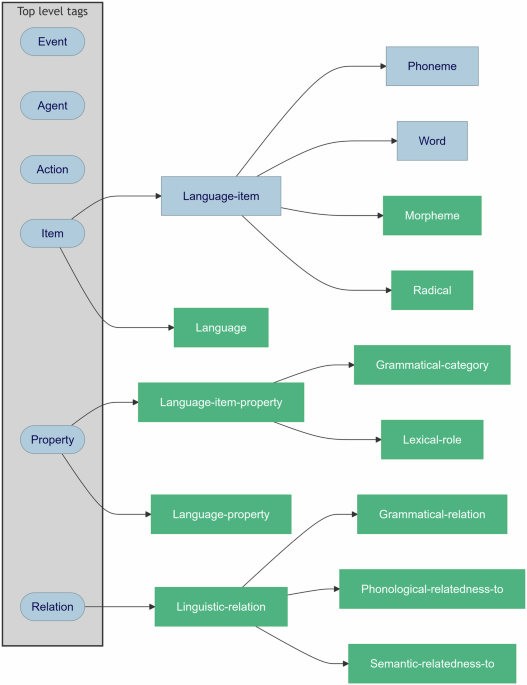
HED LANG builds on existing linguistic frameworks, incorporating terminology from these works and extending it to meet the needs of cognitive scientists. Terms were organized to fit into the existing taxonomy of the HED Standard schema, so that researchers can seamlessly use HED LANG to describe both their experimental paradigm and the linguistic properties of their stimuli. New terminology covers details about linguistic categories (e.g., part of speech, verb tense) and even properties of morphemes, allowing for fine-grained descriptions of language-related data.
HED LANG is a library extension of the HED Standard schema. Library extensions are actively supported by the HED framework to allow specific vocabularies for sub-domains. By extending the existing HED schema, HED LANG inherits all the benefits of HED while introducing new vocabulary for linguistic annotations. HED is integrated with existing (neuro)cognitive data standards such as the Brain Imaging Data Structure (BIDS) and Neurodata Without Borders (NWB). This integration means that researchers using BIDS or NWB can easily begin annotating their data with HED and HED LANG. Extensive documentation and tutorials are available here. HED also comes with a suit of tools that help with annotation and analysis. CTagger is a web application that provides a graphical user interface for data annotation. The online HED tools can be used to validate HED annotations, generate templates, and help with reorganizing BIDS event files without the need to write code using the Remodeler. All these tools help researchers quickly adopt HED LANG for their studies, making their data more shareable, discoverable, and reusable.
We tested whether HED LANG could meet the requirements of cognitive language researchers by applying annotations to recently published work in the domain language cognition. For several publications, we showed that HED LANG is able to capture the information necessary to distinguish between experimental conditions. This means HED LANG can be used to automate analysis based on standardized HED tags. Additionally we have added HED annotations to existing datasets that were shared on OpenNeuro. The annotations have been made available on the Austrian NeuroCloud.
Metadata is the backbone of FAIR data, and standards like HED LANG are essential for overcoming the challenges of annotating highly specialized research data. By adopting HED LANG, the cognitive neuroscience community can take a significant step toward FAIR data practices and more collaborative science. If you're working with language cognition data, now is the time to explore what HED LANG can do for your research!
To truly make (neuro)cognitive data FAIR, many other domain specific vocabularies are necessary. Anyone interested in extending HED can contribute directly on GitHub or contact the HED working group.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in