Encoding precision polymeric amphiphiles for direct MS decoding and label-free quantification
Published in Chemistry
The identification and biodistribution quantification of organic/polymeric nanoparticles (NPs) are essential for the evaluation of delivery efficacy and optimization of therapeutic/diagnostic functions. However, conventional fluorescent and isotopic labeling techniques rely on chemical modifications, which cannot provide actual spatiotemporal biodistribution information for soft matter NPs due to altered surface properties, uncontrolled cleavage of conjugated labels, background interference, and spontaneous/triggered degradation. Another challenge for the quantification of conventional polymeric NPs originates from inherent molecular weight distributions and undefined sequences.
Aiming to develop quantification strategies for next-generation precision NPs, we propose the following three design criteria for polymeric amphiphiles: i) appropriate mass range to bypass interferences from abundant biomolecules in a complex biological milieu; ii) high ionization efficiency to enable ideal mass spectrometry (MS) detection sensitivity under both cellular and tissue conditions; and iii) facile identification, direct sequence readout, and label-free quantification for NPs self-assembled from sequence-defined polymeric amphiphiles (digital amphiphiles) by matrix-assisted laser desorption ionization (MALDI) MS spectrometry.
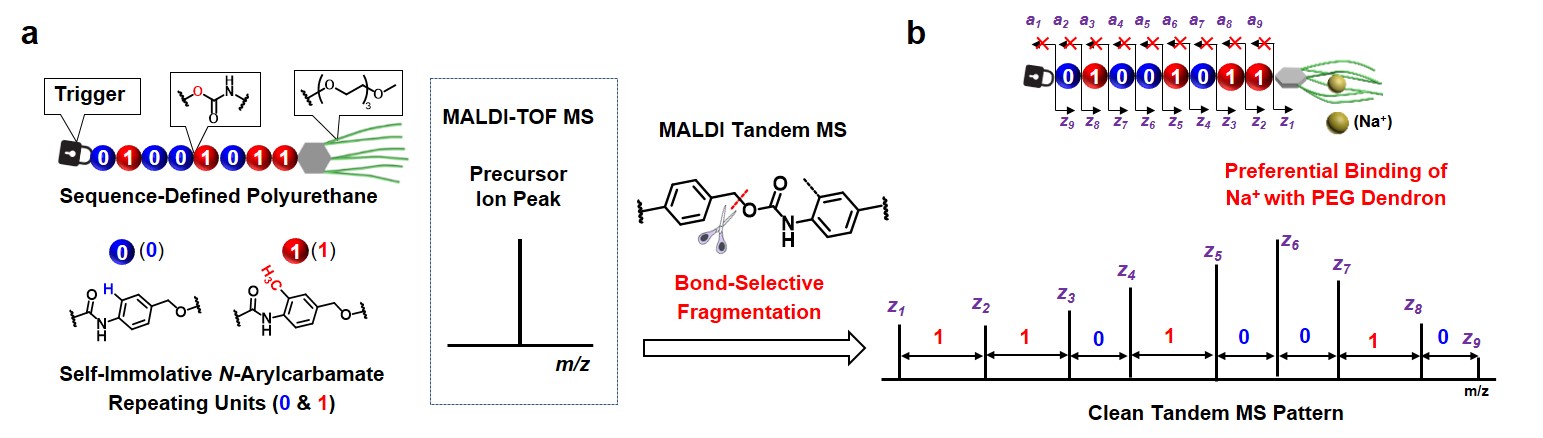
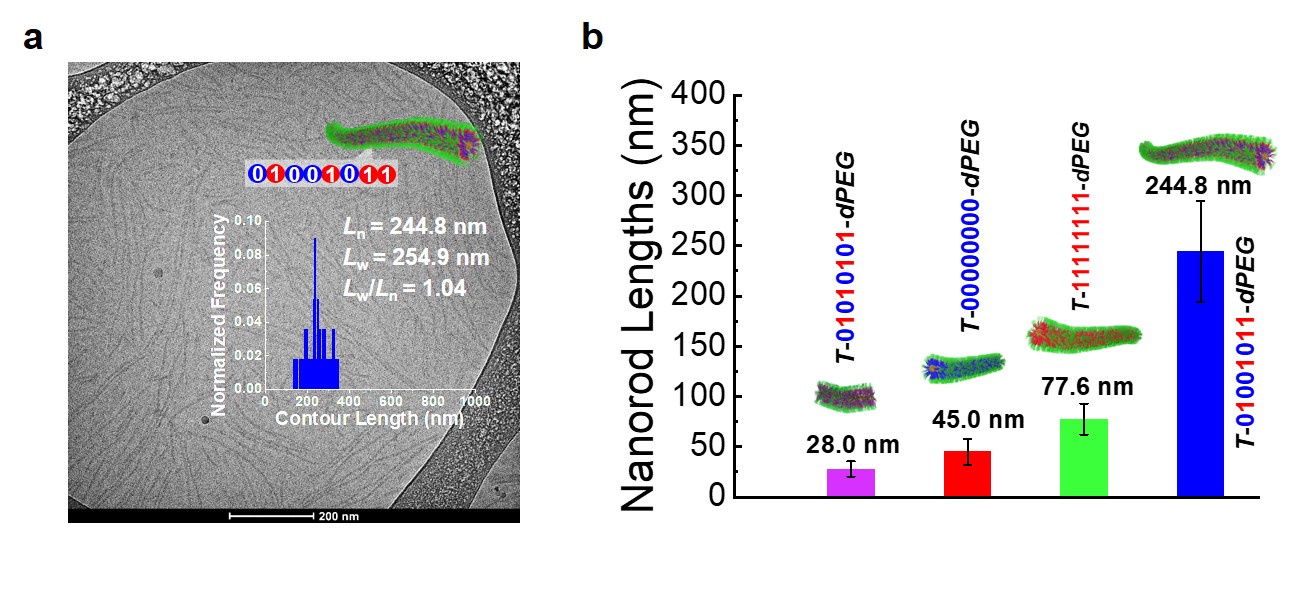
We developed a modular synthetic approach to obtain a series of encoded polymeric amphiphiles possessing light-sensitive triggers, N-aryl carbamate repeating units, and hydrophilic PEG dendrons (Fig. 1a). We discovered that the PEG dendron not only endowed amphiphilicity but also selectively bound alkali metal ions (e.g., Na+) to boost MALDI ionization efficiency and detection sensitivity. In MALDI tandem MS analysis, encoded amphiphiles revealed an unexpected clean pattern, rendering a direct sequence readout (Fig. 1b). These serendipitous results could be rationalized as follows: i) benzyl-O linkages undergo selective cleavage under MALDI tandem MS; ii) preorganization of Na+ ions onto PEG dendrons (acting as an open-chain crown ether) at the single-molecule level. In addition, these novel encoded polymeric amphiphiles are capable of self-assembling into diverse digital micelles with nanostructure morphologies and nanorod lengths strictly regulated by the lengths and primary sequences of the encoded segments (Fig. 2).
We further investigated the biodistribution of a mixture of digital micelles in animal models by taking advantage of the high-throughput nature of the MS technique. Four types of digital micelles (nanospheres and different lengths nanorods) were co-injected intravenously into SD rats, which were sacrificed and major organs are removed at different time points. The collected organs were homogenized by grinding and lyophilized. Digital micelles were extracted and quantified by MALDI-TOF MS (Fig. 3a). Furthermore, the enhanced MALDI detection sensitivity due to boosted ionization efficiency by PEG dendron (Fig. 3b) enabled MALDI imaging of tissue slices (for example, spleen slices) to reveal spatial distributions of the mixture of digital micelles (Fig. 3c). In this work, we further demonstrated the fabrication and functional applications of digital lipids and barcoded polymeric micelles.
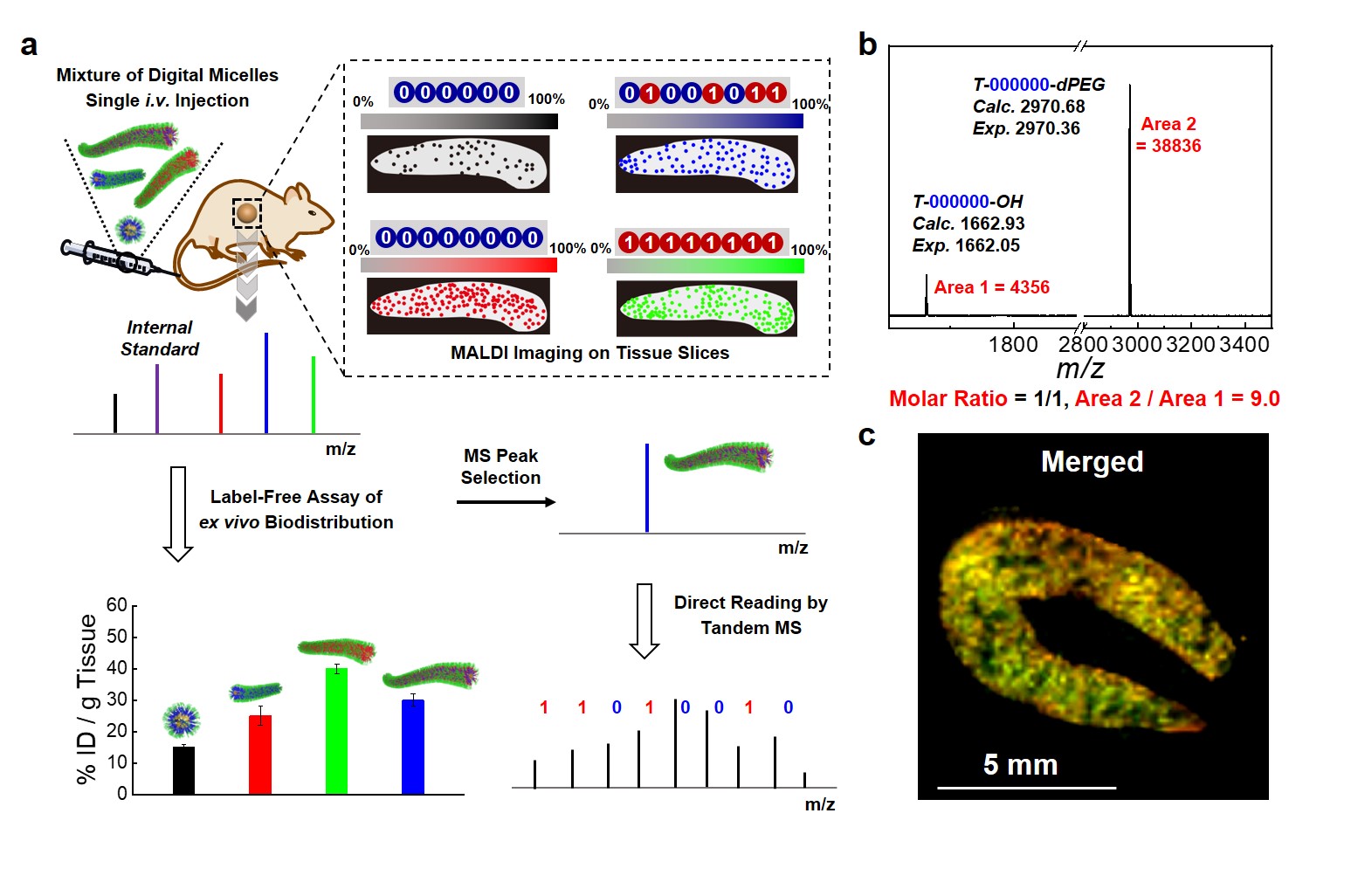
Fig. 3 Identification and label-free quantification of digital micelles at the organ and tissue slice levels. a) Upon i.v. injection of a mixture of digital micelles with varying contour lengths into SD rats, in vivo biodistribution at both the organ and tissue slice levels was assayed via MALDI-TOF MS and MALDI imaging in a label-free manner. b) MALDI-TOF MS spectrum recorded for mixtures of T-000000-OH and T-000000-dPEG at a molar ratio of 1/1, together with MS peak area ratio, demonstrating enhanced MALDI signals due to PEG-dendron-assisted ionization. c) Typical overlay MALDI imaging results of four types of digital micelles recorded for the spleen slice.
In summary, this work presents the first example of facile identification, direct reading, and label-free biodistribution quantification of digital micelles, which are self-assembled from precision synthetic polymers with built-in sequence information. We envisage that the PEG dendron-boosted MALDI detection/quantification and direct decoding strategy by MALDI tandem MS can be used to sequence other biomolecules and precision synthetic polymers and explore their chemical/biological processes and transport pathways under cellular and in vivo conditions.
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in