Engineering new-to-nature biochemical conversions by combining fermentative metabolism with respiratory modules
Published in Bioengineering & Biotechnology, Microbiology, and Mechanical Engineering

Microbial fermentation is a cornerstone of industrial bioproductions and human civilisation. The processes for making bread, alcoholic beverages or fermented milk products such as yoghurt and cheese all rely on fermentation. The term describes the cellular growth mode in absence of a terminal electron acceptor, during which the overaccumulation of reducing equivalents can only be prevented by transferring electrons to highly reduced fermentation products which are excreted by the cell. Since maintaining the cellular redox balance is essential for cell growth, fermentatively growing cells are inherently forced to secrete the majority of substrate taken up as fermentation products into the medium. This redox-balancing requires precise stoichiometric matching of electrons taken up and secreted. Therefore, only certain substrate-product combinations allow such matching (are thus redox-balanced), and thus allow fermentative cell growth. For example, anaerobically grown E. coli cells will typically ferment glucose to various products including lactate, acetate, or ethanol (heterofermentative growth) while they cannot grow on reduced carbon sources like glycerol.
To overcome this limitation, we envisioned a combination of respiratory and fermentative growth. Notably, these two growth modes naturally occur in distinct environments and are tightly controlled by highly conserved and ancient regulatory systems. Thus, we first wondered whether we could engineer the transition from respiratory to fermentative growth in aerobic conditions. Here, we decided to enforce the fermentative growth mode by manually deleting any route allowing electron transfer to the respiratory chain via reduced quinones. Both literature search and computational predictions allowed us to identify knockout targets which would prohibit quinone reduction. Over the time course of six years, multiple authors then constructed the obligate fermenting NNmini strain carrying 19 gene knockouts of NAD(P)H dehydrogenases, quinone oxidoreductases and "mini-cycles", small metabolic cycles transferring electrons from NADH to quinones (Fig. 1B). Upon characterising the strain, we found that it indeed aerobically converted supplied glucose stoichiometrically and exclusively to lactate (Fig. 1C), and accumulated reducing equivalents to an extend that is consistent with anaerobically grown cells.
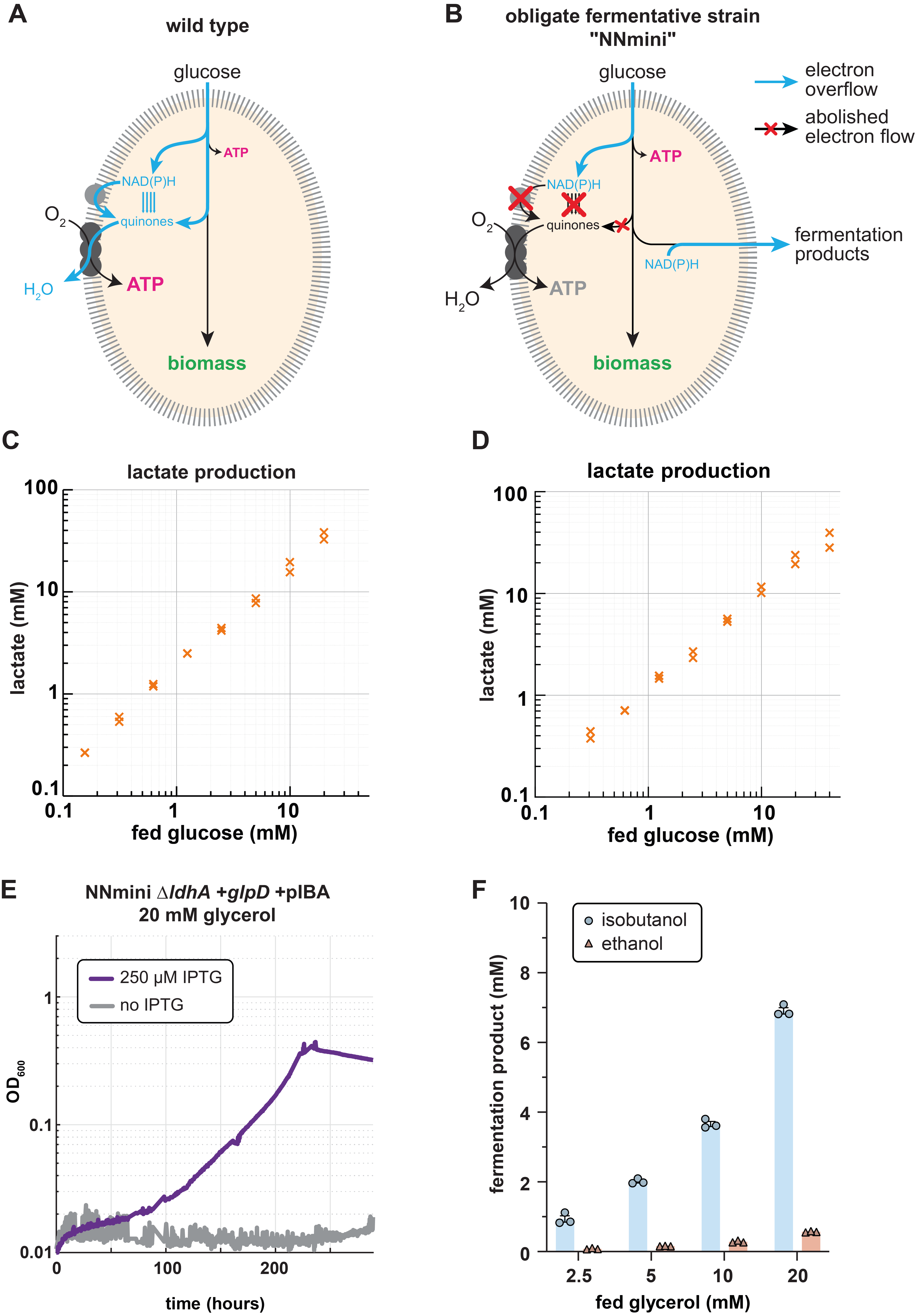
After confirming the fermentative phenotype of the NNmini strain, we sought to engineer the fermentation of novel substrate-product combinations. Canonically, fermenting glycerol to lactate results in redox imbalance (in E. coli) since one NADH is formed that cannot be recycled through lactate formation. If, however, a quinone instead of an NADH is formed, the process would be redox-balanced if the quinone can be used to transfer the electron pair to a terminal electron acceptor. Therefore, as first demonstration, we added the quinone dependent glycerol 3-phosphate dehydrogenase (GlpD) to reduce the amount of NADH that is generated, which should result in a redox-balanced process. To reduce the engineering effort for adding the previously deleted glpD to the NNmini strain, we made a P1 lysate from an engineering intermediate that still contained glpD and transduced the NNmini strain with it. When we plated on minimal medium plates containing only glycerol after the transduction, all occurring colonies contained the glpD locus again. Upon confirming that indeed only glpD had been reintroduced, we further could demonstrate glycerol fermentation to lactate by the NNmini +glpD strain (Fig. 1D). This first success in engineering new substrate-product combinations encouraged us to further push the metabolic boundaries of our strain by directing the fermentative flux towards isobutanol instead of lactate production. Indeed, upon overexpressing the isobutanol synthesis pathway and deleting lactate dehydrogenase, we found that our strain was able to selectively produce isobutanol from glycerol for the first time (Fig. 1E, F).
In summary, we constructed the a respiro-fermentative strain as platform for novel fermentations that can employ respiratory modules for highly selective electron transfer to the respiratory chain. Respiration and fermentation as two highly conserved growth modes are combined to open up novel avenues for microbial bioproductions. The engineering endeavour was not only complex for the challenging metabolic rewiring, but also survived the transition of all authors to other institutes due to the sudden death of the group leader Dr. Arren Bar-Even. Dr. Bar-Even pioneered bold metabolic engineering and was an inspirational teacher to everyone involved in this study. He is sorely missed. A multi-institute collaboration of all authors made it possible to wrap up this work and hopefully pave the way for new bioproductions in the future.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in