Enhancing Clinical Mutation Detection: A Game-Changing Molecular Approach
Published in Genetics & Genomics
Molecular diagnostics in cancer research has underscored the pivotal role of driver genetic mutations in tumor growth and metastasis. The advent of liquid biopsies, which involve the detection and quantification of genomic alterations in circulating cell-free DNA (cfDNA) from blood samples, has introduced a paradigm shift in the field. These liquid biopsies offer unmatched advantages, including convenience, minimal invasiveness, and heightened sensitivity for early-stage tumor detection.
However, a significant hurdle remains in the detection of mutations, as they often exist at extremely low frequencies (<0.1%). Current mutation detection methods can be broadly categorized into direct detection techniques, like quantitative polymerase chain reaction (qPCR), droplet digital PCR (ddPCR), and sequencing, and enrichment-based methods such as BDA, NaME-PrO, DASH, and NAVIGATER. Yet, these methods face limitations concerning detection limits, complex probe designs, expensive instrumentation, and susceptibility to false positives.
In this context, we introduce a revolutionary DNA-excision method for the detection of low-frequency mutations, both in genomic DNA and cfDNA, at the single-nucleotide level. This breakthrough is rooted in a competitive DNA-binding-and-digestion mechanism guided by single-stranded phosphorothioated DNA (sgDNase), which effectively removes wild-type DNA strands (Fig. 1). Unlike traditional applications of deoxyribonuclease I (DNase) as a non-specific endonuclease, our method leverages single-stranded phosphorothioated DNA's (sspDNA) superior binding affinity to DNase. This unique feature guides DNase specificity through competitive binding and digestion, enhancing single-nucleotide resolution.
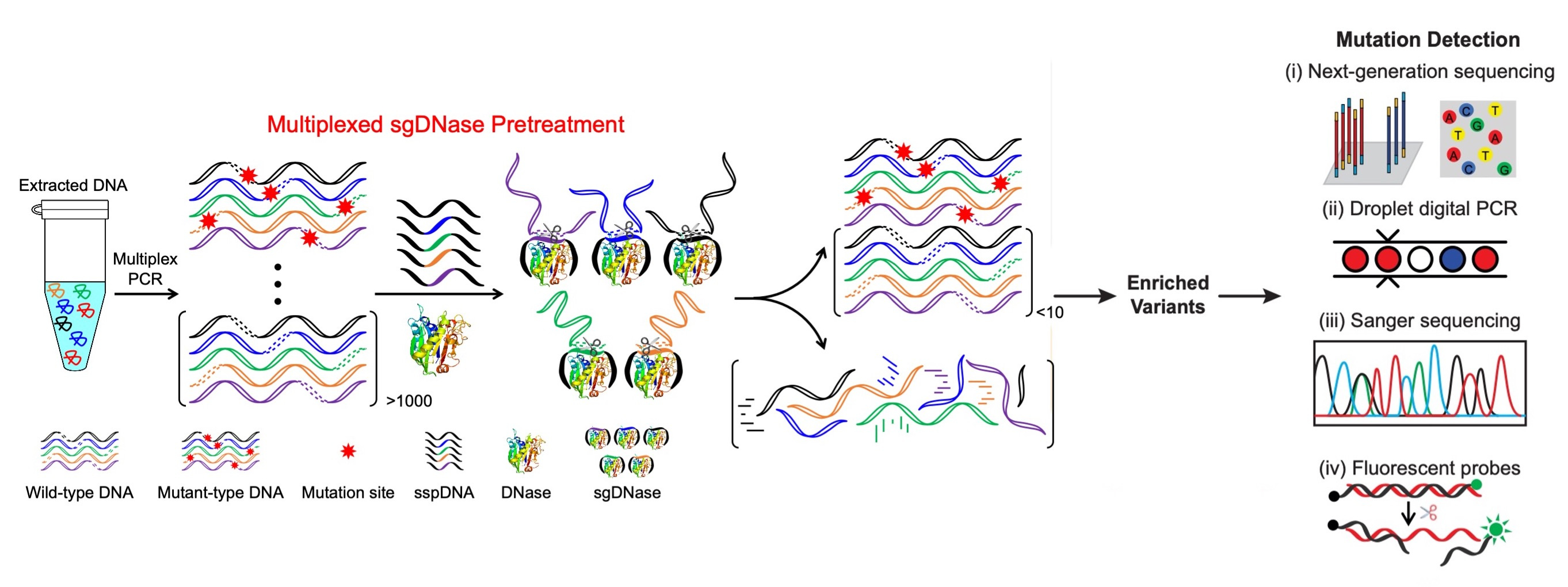
Fig. 1 Illustration of the sgDNase system for multiplexed enrichment of clinically relevant target mutations via a competitive binding and digestion mechanism.
Proof of concept was established through the detection of the EGFR L858R mutation, a critical driver gene mutation in non-small cell lung cancer. Our sgDNase system, guided by S1-EGFR sspDNA, demonstrated exceptional single-nucleotide discrimination capabilities, achieving a discrimination factor (DF) of 166. The single-nucleotide resolution was confirmed through High-Performance Liquid Chromatography (HPLC)(Fig. 2a).
Taking advantage of sgDNase's single-nucleotide resolution, we devised the "sgDNase pretreatment" platform to enrich low-level mutant strands for subsequent mutation detection methods such as NGS, ddPCR, Sanger sequencing, or fluorescent-probe-based assays. Remarkably, sgDNase pretreatment elevated variant allele frequencies (VAFs) from minimal levels to substantially higher percentages across various target sequences, demonstrating its universal applicability(Fig. 2b).
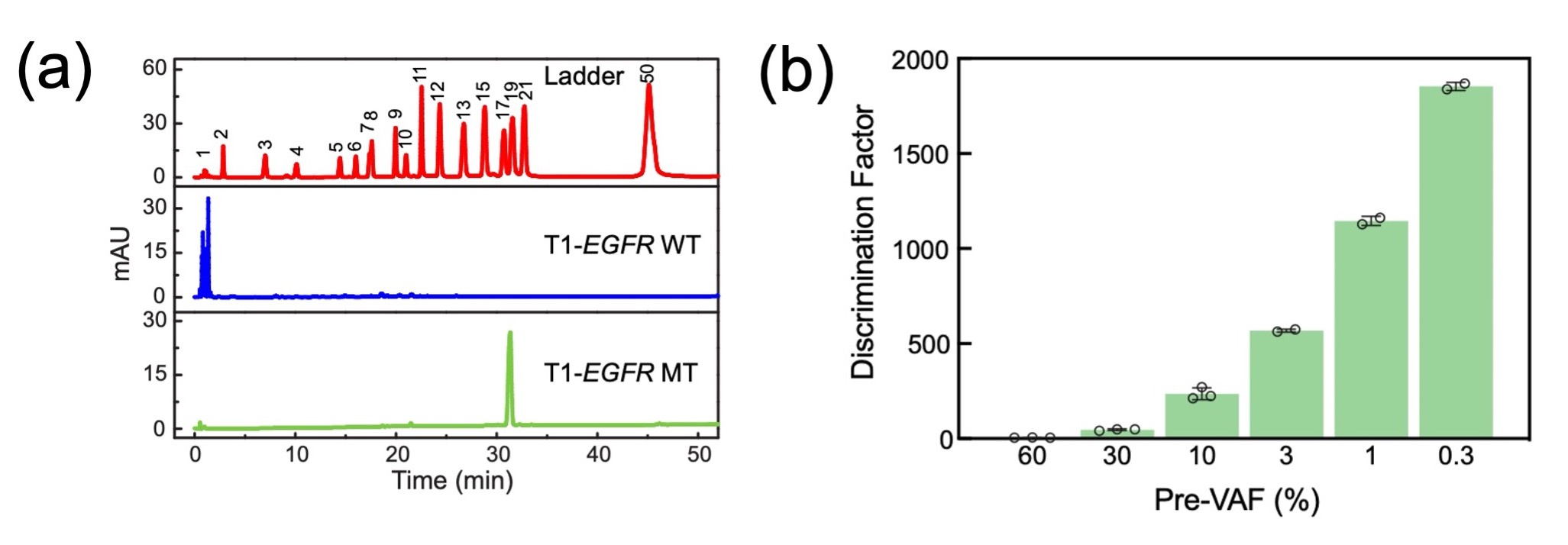
Fig. 2 (a) HPLC analysis of the digestion products of unlabelled EGFR L858R WT and MT strands by sgDNase. (b) Relative discrimination efficiency (DE) of sgDNase in the cleavage of P1-EGFR WT-Cy5 and P1-EGFR MT strands at different VAFs (60–0.3%). DE represents the ratio of the digestion rate of WT to that of MT in mixed substrates.
Furthermore, sgDNase's versatility shone through in scenarios with highly variable mutation types within mutational hotspots (Fig. 3a). It effectively enriched all different types of mutations and combinations within the region, opening the door to the rapid identification of rare and unknown mutations. We extended the application to pretreat human genomic DNA (dsgDNase) directly. Clinical samples' genomic DNA underwent a straightforward process, resulting in notable VAF increases for the EGFR L858R mutation. This method's reliability was confirmed through comparison with results from ddPCR and Sanger sequencing, validating its efficiency in detecting mutations from low VAFs.
The application of the sgDNase pretreatment method to detect ultra-low-abundance mutations in clinical biopsy samples yielded significant findings. Out of more than 70 clinical samples analyzed, a total of 16 positive samples were identified. Before employing sgDNase pretreatment, only two tissue samples (Patients P2 and P5) were found to be positive for the EGFR L858R mutation, with no positive blood samples detected. However, following sgDNase pretreatment, the mutation was detected in an additional tissue sample (Patient P7) and two blood samples (Patients P2 and P10), as shown in Fig. 3b. These positive findings were further confirmed through Next-Generation Sequencing (NGS). In the case of KRAS G12/G13 mutations, blood sample P14-B was identified as KRAS G13D positive after pretreatment, aligning with the tissue sample results (Fig. 3c). Most remarkably, sgDNase pretreatment uncovered the presence of KRAS G13S mutation in both the blood and tissue samples of Patient P14, indicating a double-positive status (KRAS G13D + G13S). Furthermore, blood sample P15-B and tissue sample P18-T were both identified as KRAS G12V mutation positive after undergoing sgDNase pretreatment, highlighting the method's capability to uncover crucial genetic insights in clinical biopsy samples.
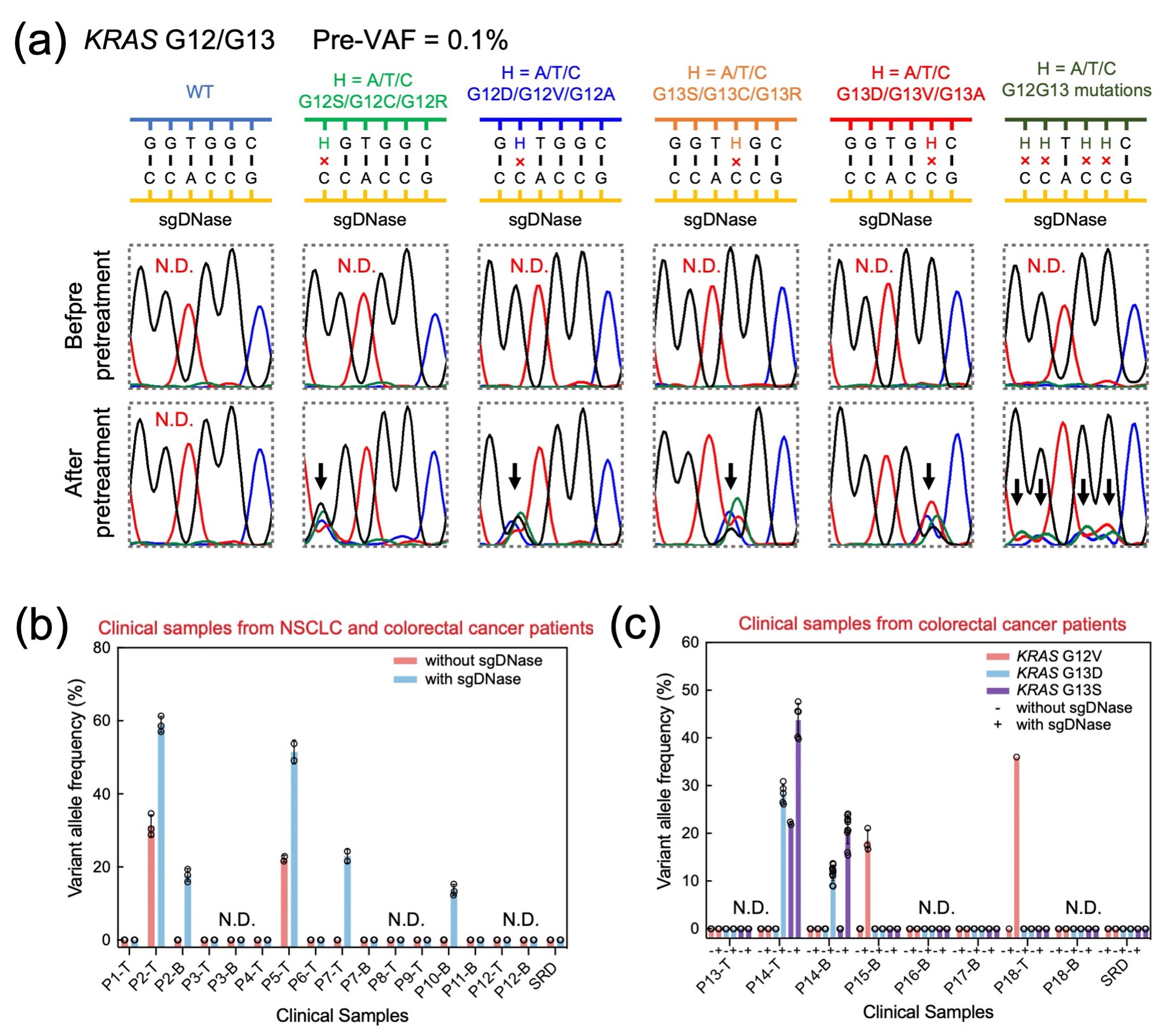
Fig. 3 (a) Comparison of Sanger sequencing chromatograms of synthetic KRASG12G13 samples (flora text for G12S, G12C and G12R; blueberry text for G12D,G12V and G12A; tangerine text for G13S, G13C and G13R; maraschino text for G13D, G13V and G13A; and moss for their combinations) with pre-VAF at 0.1% before and after sgDNase pretreatment. The curves in different colours represent different nucleotides (blue for C, black for G, red for T and green for A). The elevated mutation level after pretreatment is indicated by a black arrow above the corresponding chromatographic peak. (b-c) Detection of ultra-low-abundance mutations in clinical biopsy samples with sgDNase pretreatment. (b) sgDNase applied to 16 cfDNA and gDNA samples from eight NSCLC patients (P1–P8), four colorectal cancer patients (P9–P12) and standard reference DNA (SRD). P, T and B in the sample name represent patient, tissue sample (gDNA) and blood sample (cfDNA), respectively. (c) Rare mutation identification in KRAS G12/G13 residues in eight cfDNA and gDNA samples from six colorectal patients (P13–P18) and SRD using sgDNase-Sanger sequencing.
In summary, sgDNase presents a powerful DNA excision tool capable of specifically digesting unwanted DNA sequences with single-nucleotide precision. This innovative approach eliminates wild-type sequences while enriching mutant strands. The potential applications of sgDNase are vast, ranging from continuous mutation status assessment to enhancing the prospects of precise cancer therapy.
The breakthrough described here exemplifies how a smart molecular recognition reaction can outperform large-scale, expensive sequencing methods, accelerating detection, and cost savings. It holds immense promise for advancing clinical mutation detection capabilities, driving precision medicine, and improving treatment outcomes.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in