
It started with a thirty-minute meeting I had in 2014 with my PhD supervisors and now corresponding authors, Tom de Greef and Luc Brunsveld. We discussed that we could combine the experience I gathered during an internship for my Master's degree with our lab's knowledge on enzymes involved in programmed cell death. A nice and easy project to start my PhD with—we thought naively.
What followed was a scientific adventure that spanned five years and almost my entire PhD. Looking back on all the challenges and setbacks we encountered along the way, we could have (and maybe should have) pulled the plug on the project several times. I am happy we didn't.
The idea behind the paper was simple. Our lab had experience with caspase-9, a key enzyme in one of the apoptotic pathways. It becomes active upon dimerization when recruited to the apoptosome, a multivalent scaffold in the cell. We wanted to create a modular platform on which we could investigate the molecular interactions between caspase-9 monomers in systematic fashion—a true synthetic biology approach. Is a couple of caspases enough? And if two's a company, is three or four a crowd? And does caspase catalysis require one active site or two?
The nanoscale platform was constructed using the DNA origami technique, which I was introduced to during my internship in the lab of William Shih, a pioneer and expert in the field. The DNA I ordered arrived in a big box just before the holidays in 2014, and I folded—well, tried to fold—my first batch of DNA origami soon after.
Optimization of the DNA origami assembly proceeded relatively quickly. It was the next step in the project that almost made us give up on the whole thing.
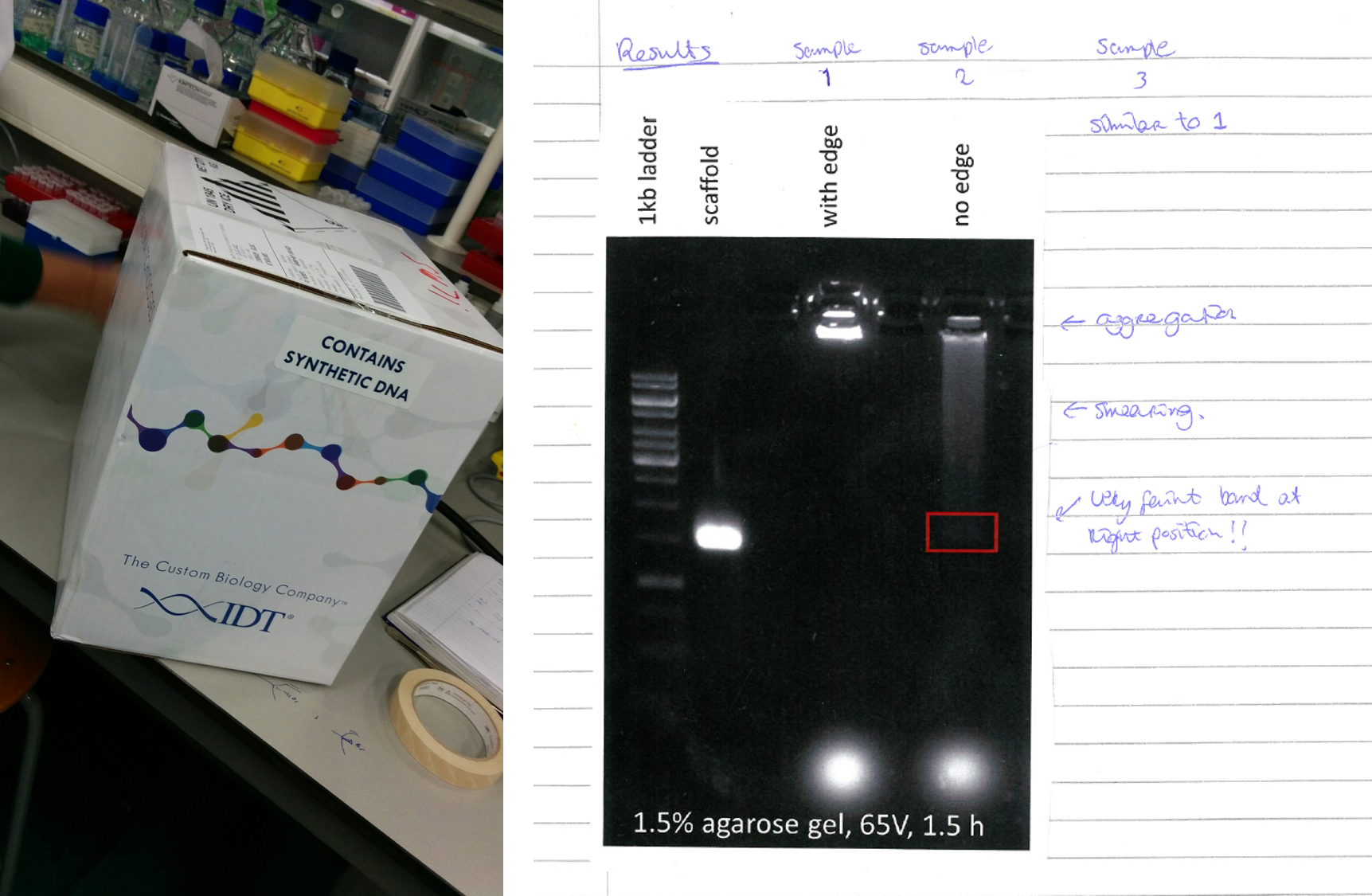
The synthesis of the caspase-9 enzyme-DNA conjugates was difficult. Very difficult. It took me more than two years to get the job done, luckily with help from co-author and caspase connaisseur Anniek den Hamer. The funny thing is that this frustratingly long process ended up in Figure 1d with a simple two-lane gel: enzyme before and after conjugation. Period.
Along with the difficult moments in the project, I vividly recall moments of pure happiness. I remember seeing that first band indicating successful conjugation after staring intensely at a PAGE gel destaining at 11 at night. I remember Berta Gumí Audenis meticulously brute-forcing her way through an endless list of AFM measurements, and that meeting with model gurus Bart Markvoort and Job Roodhuizen where we realized that the experiments of Figure 4 matched with the models. I also remember me fist pumping while doing laps around the lab as soon as I saw the platereader producing the results for Figure 5.
In the end, we actually found the answer to more questions than we had at the start of the project. It also made us firmly believe that platforms based on folded DNA can be applied to unfold details of other enzymes. Examples could be caspase-8, caspase-1, the IRAK kinases, and the kinases and phosphatases in Ste5-based signaling.
As long as you get that tricky enzyme-DNA conjugation under control.
Follow the Topic
-
Nature Catalysis

This journal brings together researchers from across all chemistry and related fields, publishing work on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, incorporating both fundamental and applied studies.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in