Evolutionary patterns of CXCL16-CXCR6: a deep dive into vertebrate lineages
Published in Ecology & Evolution, Genetics & Genomics, and Immunology
CXCR6 deficiency lowers CD8 tissue-resident memory (TRM) cell numbers in the lungs and depletes ILC3s in the lamina propria, impairing mucosal defence [1, 2]. We explored the evolution and diversification of the CXCL16-CXCR6 axis in more than 400 vertebrate genomes spanning fishes, amphibians, reptiles, birds, and mammals. Through a comparative genomic analysis, we discovered that the unique DRF motif varies across the vertebrate lineages and recurrent and concurrent loss of these genes in 10 out of 36 bird orders (such as Galliformes, Passeriformes, and others), Crocodylia, and Elapidae snakes.
Studying CXCR6 protein orthologs across vertebrates allows us to uncover variations in the DRF motif and how it impacts G-protein interactions.
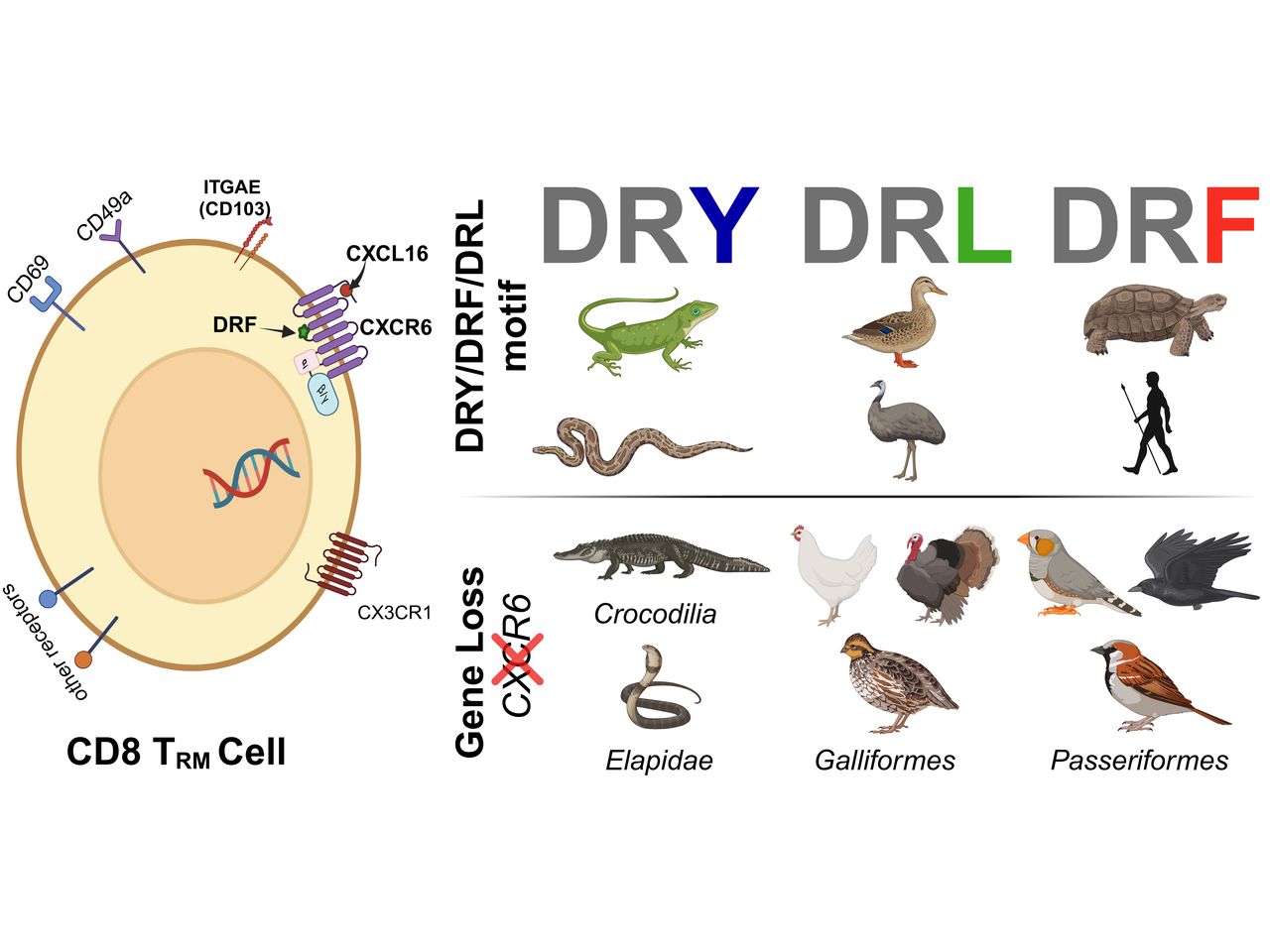
The DRF motif in CXCR6 interacts with Gi/o proteins and is an adaptation to adhesion [3, 4]. By performing evolutionary trace analysis, we identified that this motif exhibits diversity across vertebrate lineages. Mammals, turtles, and frogs primarily exhibit the DRF motif, while birds typically display a DRL variant, except for the harpy eagle, which possesses the DRF motif. Snakes and lizards, in contrast, possess the more common DRY motif, characteristic of many chemokine receptors (CKRs) (Figure 1).
Our analysis revealed convergent amino acid substitutions in both Aves and Squamates, particularly at the DRF motif and two other key positions. This convergence suggests that evolutionary pressures may have driven similar molecular adaptations in these distinct lineages. To explore the functional implications of these motif variations, we used molecular dynamics simulations, which showed that changes at the DRY/DRF/DRL site influence the interaction between CXCR6 and G-proteins. Specifically, the DRL variant found in birds demonstrated a reduced coupling probability with the GoA G-protein, as predicted by the machine learning tool PRECOGx [5]. This reduced interaction may suggest a different regulatory mechanism of G-protein signalling in birds compared to other vertebrates. Furthermore, our findings can inform future functional studies, as previous research has used receptors like CCR6 [4] and CX3CR1 [3] to investigate the importance of the DRF motif in CXCR6. Further studies utilizing CXCR6 orthologs from various species with heterogeneity at the DRF motif will offer deeper insights into the evolutionary and functional significance of the DRY/DRF/DRL motif.
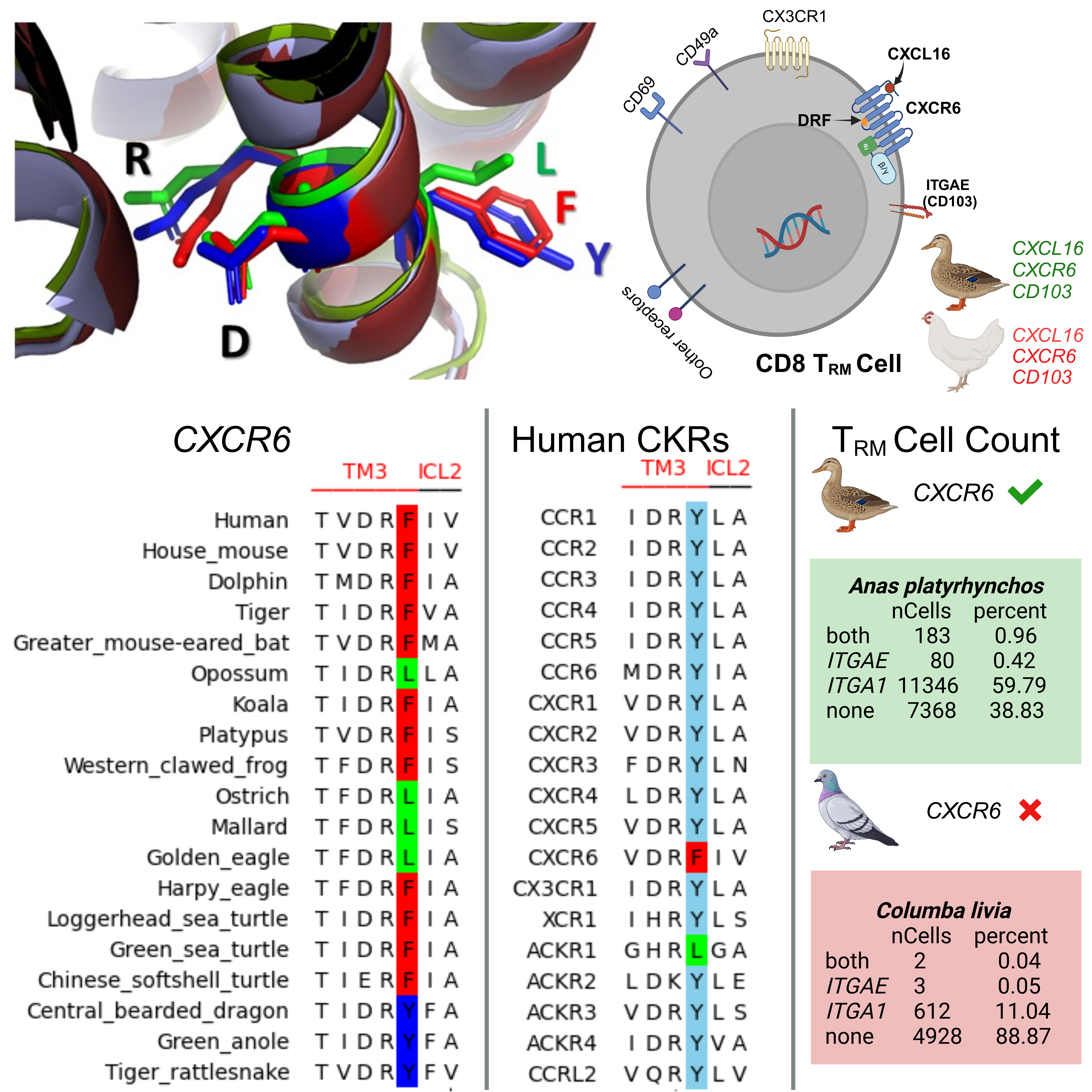
Figure 1: The top-left panel highlights the variation in the DRF motif (DRY/DRF/DRL) within the H3C region across vertebrate species (left column) compared to the conserved DRY motif for other human chemokine receptors (middle column). The top-right panel illustrates a CD8 TRM cell with its marker genes. The third column compares the impact of CXCR6 gene loss on CD8 TRM cell count (the gene is intact in Mallard and lost in Pigeon). TRM cells were identified using markers genes ITGAE and ITGA1.
Recurrent and concurrent loss of CD8 TRM cell-related genes: CXCL16, CXCR6, and ITGAE
Our comparative study revealed a recurrent loss of the CXCR6 gene in 10 out of 36 bird orders as well as in Crocodilia and the Elapidae family of snakes. This gene loss results from segmental deletions and/or frame-disrupting changes. The functional consequences of CXCR6 loss have been documented in mice, where its absence leads to a reduction in CD8 TRM cells in the lungs [1,2]. We observed a similar trend in birds: species lacking CXCR6 showed a decrease in TRM cell numbers, as evidenced by our analysis of limited single-cell RNA sequencing data of lung tissue from two bird species—mallard, which retains intact CXCR6, and pigeon, which has lost this gene. (Figure 1). The concurrent loss of CXCR6 and ITGAE (CD103) genes in chickens raises important questions about our definitions and understanding of TRM cells in avian species (Figure 2).
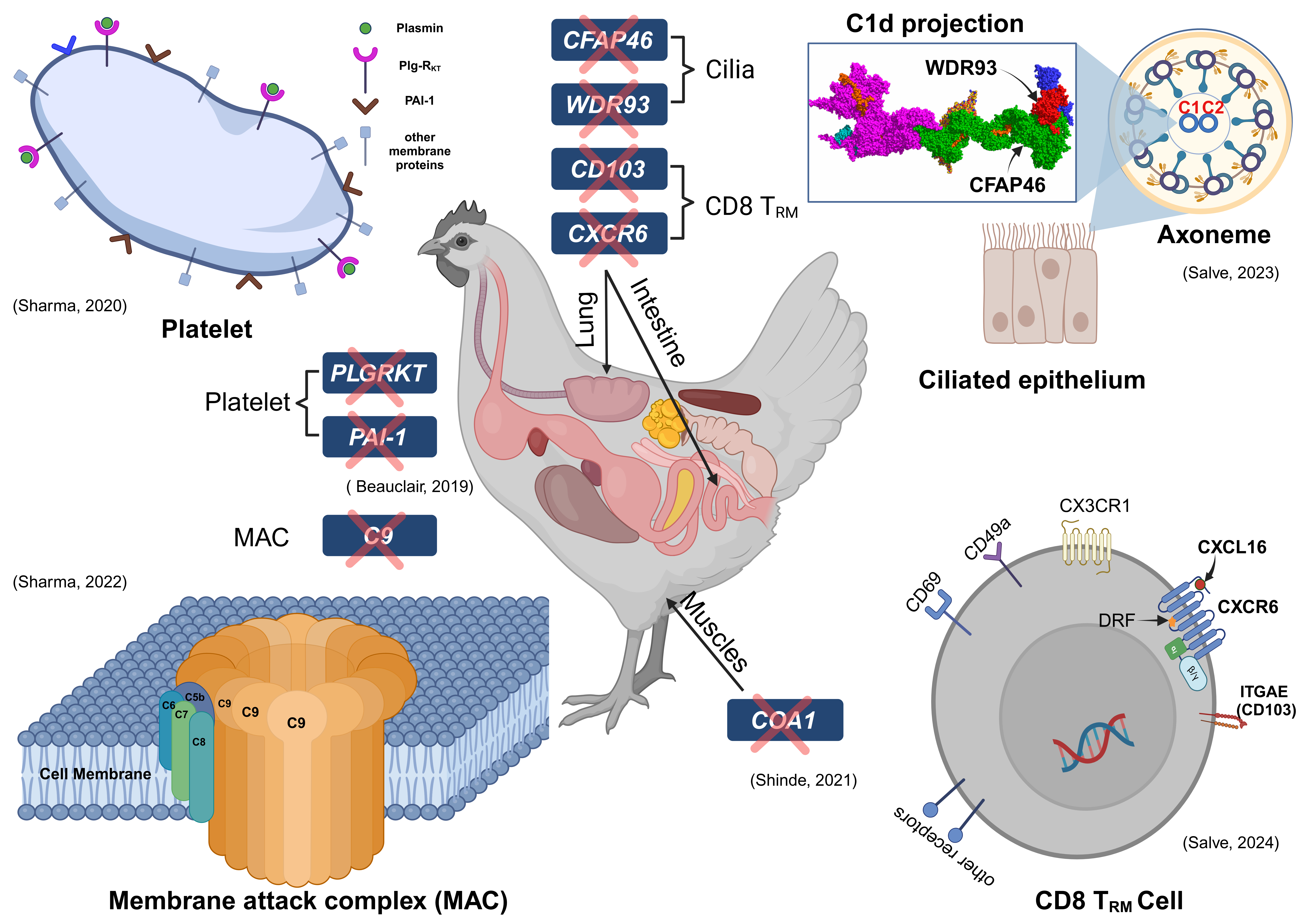
Figure 2: Illustration of organs, anatomical sites, and cells in chicken where the lost genes play an important role.
Impact of gene loss: Differential susceptibility to avian influenza virus
Considering the important role of the CXCL16-CXCR6 axis in the innate-adaptive immune system, comparative genomic analyses focusing on gene presence/absence patterns and adaptive evolution enable an understanding of the link between inter-species genetic differences and disease susceptibility. Moreover, CXCR6 serves as a core signature gene for CD8 TRM cells, alongside other genes such as CD69, ITGAE (CD103), and ITGA1 (CD49a) [1]. These tissue-resident memory (TRM) cells are essential for mounting effective immunity against influenza infection, for example In CXCR6-/- mice, compromised cellular immunity to influenza infection has been observed [1]. In our previous study titled “Concurrent loss of ciliary genes WDR93 and CFAP46 in phylogenetically distant birds” (DOI: https://doi.org/10.1098/rsos.230801), we reported the loss of ciliary genes WDR93 and CFAP46 in geese and Galliformes. Birds lacking these genes, primarily from the superorder Galloanserae, show varied susceptibility to avian influenza virus (AIV), with geese being tolerant carriers and galliform birds highly susceptible. In contrast, ducks retain transcriptionally active, intact genes and are resistant to most AIV strains. For more details on this work, please watch the presentation by Buddhabhushan at the Evolution 2024: virtual conference.
Together, this highlights that lineage-specific gene losses may lead to differential susceptibility to avian influenza virus. The loss of CFAP46, WDR93, ITGAE, CXCL16, and CXCR6 in chickens may have altered CD8 TRM cell abundance and cilia functioning, with implications for immunity against viral diseases and vaccines inducing CD8 TRM cells (Figure. 2). While linking gene loss to its phenotypic effect, the gene knockout and gene knockdown studies are greatly helpful. For instance, the difference in cilia functioning can be seen in an inter-species comparison study of birds, in which species-ostrich, which has an intact WDR93 and CFAP46, and turkey, which has lost these genes [6]. However, other bird species remain completely unstudied. So, future studies can be done to check how cilia function without these genes. Similarly, we used single-cell transcriptomic data for the lungs of two bird species—mallard, which has an intact CXCR6, and pigeon, which has lost CXCR6, to check the effect of gene loss TRM cell counts. CXCR6 deficiency in mice leads to impaired cancer vaccine efficiency [7]. Consequently, species with CXCR6 gene loss could be targeted for improved immunotherapies and vaccination strategies.
Our study opens new avenues for understanding how gene repertoires vary across species and underscores the importance of evolutionary comparative genomics in studying immune functions across vertebrates.
Understanding the Evolutionary Patterns: Concurrent and Recurrent Gene Loss
Previously, we reported the concurrent loss of ciliary genes CFAP46 and WDR93, in phylogenetically independent birds [8]. This raises an intriguing question: which gene was lost first, or is it a classic chicken-and-egg scenario? Previous research indicates that the CFAP46 mutant negatively impacts WDR93, preventing the assembly of the c1d projection [9]. Given the substantial size of the CFAP46 gene region approximately 90 kb. It is plausible that this gene was lost first due to segmental deletion, leading to the loss (via frame-disrupting changes and/or substitutions leading to in-frame STOP codon) of its interacting partner, WDR93. Similarly, the CXCL16-CXCR6 axis presents another interesting case of potential concurrent and recurrent gene loss, particularly within sauropsids [10]. Here, a possible explanation points to CXCL16, which shows an increase in GC content, making it more prone to loss [11]. In sauropsids (including Aves), where the GC content exceeds 65%, CXCL16 interacts exclusively with CXCR6, suggesting that both genes (ligand-receptor axis) may have been lost concurrently due to their interdependence. These examples illustrate the concepts of recurrent loss likely driven by high GC content and concurrent loss of CXCL16-CXCR6 stemming from exclusive interactions between genes. Species that retain these genes may have functional advantages, underscoring the importance of gene integrity. While we can identify these evolutionary patterns, the absolute mechanisms behind them remain elusive.
In summary, our findings provide a detailed understanding of gene loss mechanisms, including frame-disrupting changes, segmental deletions, promoter deletions, and evidence of RELAX selection. We also computationally demonstrate how variations in the DRF motif affect G-protein selectivity. Using the GC-rich gene CXCL16 as a case study, we reveal that some genes may appear lost but are “missing” from the genome assembly. This insight paves the way for further research into similar cases. Additionally, we highlight the presence of segmental deletions and present a workflow that can be followed in future studies to confirm these deletions.
References
- Wein AN et al. 2019 CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med. 216, 2748–2762. (doi:10.1084/JEM.20181308)
- Satoh-Takayama N, Serafini N, Verrier T, Rekiki A, Renauld JC, Frankel G, DiSanto JP. 2014 The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity 41, 776–788. (doi:10.1016/J.IMMUNI.2014.10.007)
- Koenen A, Babendreyer A, Schumacher J, Pasqualon T, Schwarz N, Seifert A, Deupi X, Ludwig A, Dreymueller D. 2017 The DRF motif of CXCR6 as chemokine receptor adaptation to adhesion. PLoS One 12, e0173486. (doi:10.1371/JOURNAL.PONE.0173486)
- Singh SP, Foley JF, Zhang HH, Hurt DE, Richards JL, Smith CS, Liao F, Farber JM. 2015 Selectivity in the use of Gi/o proteins is determined by the DRF motif in CXCR6 and Is Cell-type specifics. Mol. Pharmacol. 88, 894–910. (doi:10.1124/mol.115.099960)
- Matic M et al. 2022 PRECOGx: exploring GPCR signaling mechanisms with deep protein representations. Nucleic Acids Res. 50, W598–W610. (doi:10.1093/NAR/GKAC426)
- Burn A, Schneiter M, Ryser M, Gehr P, Rička J, Frenz M. 2022 A quantitative interspecies comparison of the respiratory mucociliary clearance mechanism. Eur. Biophys. J. 51, 51. (doi:10.1007/S00249-021-01584-8)
- Karaki S et al. 2021 CXCR6 deficiency impairs cancer vaccine efficacy and CD8 + resident memory T-cell recruitment in head and neck and lung tumors. J. Immunother. Cancer 9, 1–12. (doi:10.1136/jitc-2020-001948)
- Salve BG, Kurian AM, Vijay N. 2023 Concurrent loss of ciliary genes WDR93 and CFAP46 in phylogenetically distant birds. R. Soc. Open Sci. 10. (doi:10.1098/RSOS.230801)
- Brown JM, DiPetrillo CG, Smith EF, Witman GB. 2012 A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell Sci. 125, 3904–3913. (doi:10.1242/jcs.107151)
- Salve BG, Sharma S, Vijay N. 2024 Evolutionary diversity of CXCL16-CXCR6: Convergent substitutions and recurrent gene loss in sauropsids. Immunogenet. 2024 , 1–19. (doi:10.1007/S00251-024-01357-5)
- Hara Y, Kuraku S. 2023 The impact of local genomic properties on the evolutionary fate of genes. Elife 12. (doi:10.7554/eLife.82290)
Follow the Topic
Ask the Editor - Immunology, Pathogenesis, Inflammation and Innate Immunity
Got a question for the editor about the complement system in health and disease? Ask it here!
Continue reading announcement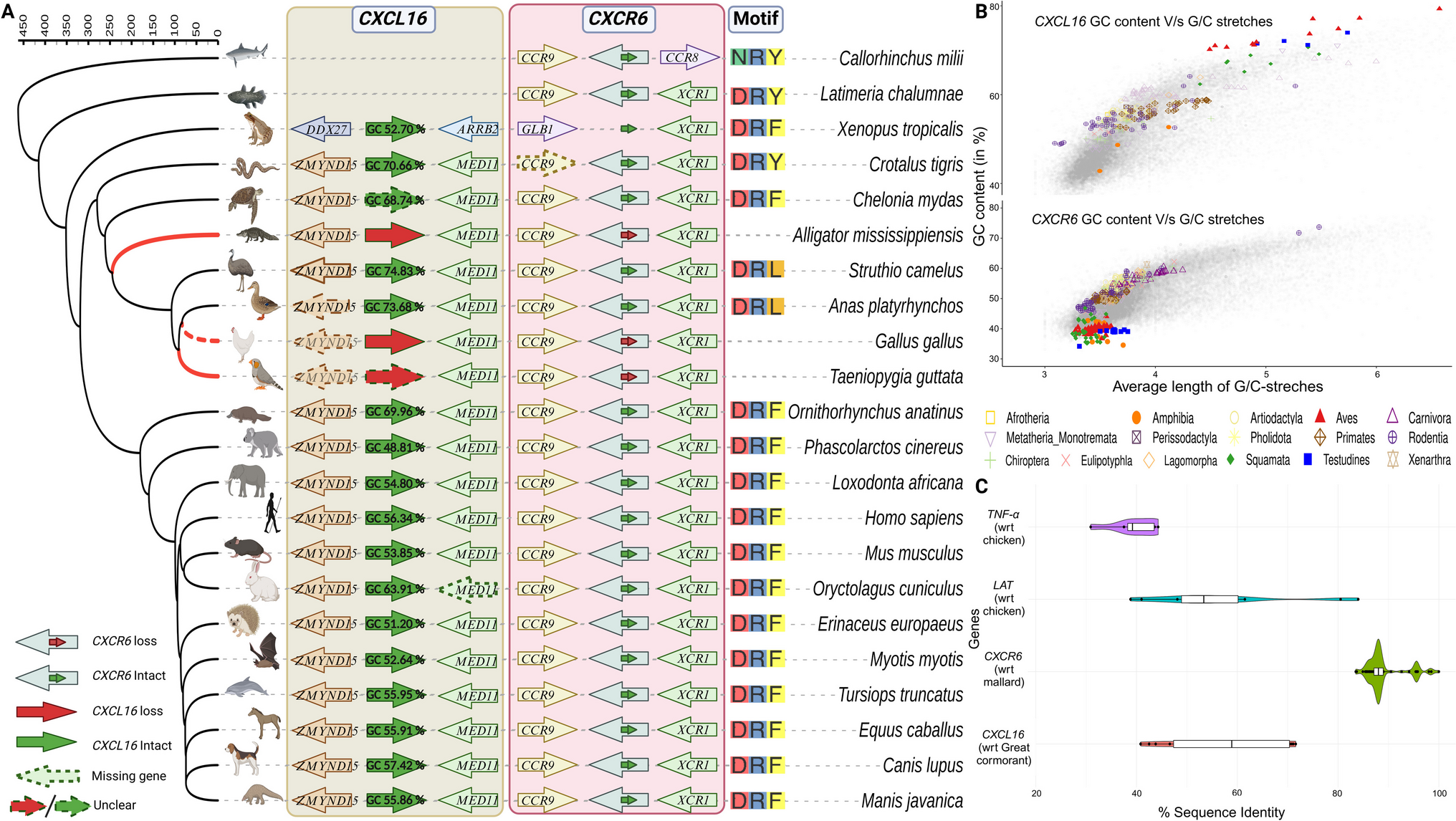



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in