Exploration of metagenome-assembled genomes sheds light on enigmatic mesophilic, marine archaea
Published in Microbiology

The Tara Oceans “-omics” datasets (Alberti et al. 2017) have reshaped the way that microbial ecologists are able to interact with the organisms that inhabit the world’s oceans. The open nature of the data allows it to live beyond the bounds originally envisioned for it by the consortium of scientists who conceived and executed the data generation. One such avenue has been the large-scale generation of large of metagenome-assembled genomes (MAGs) through our work (Tully, Graham, & Heidelberg 2018) and others (Delmont et al. 2018 & Parks et al. 2017). In the initial phase of working with our MAGs, I noticed a large number of genomes that could be assigned to the Marine Group II Euryarchaea (MGII), an enigmatic group of mesophilic archaea that had no cultured isolates and a limited amount of genomic information. It seemed like a prime opportunity to formally expand on our understanding of a phylogenetically-diverse, abundant, and globally-distributed group of organisms.
But like many good ideas in science, I was not the only one who had it. Chris Rinke and others at the University of Queensland (Rinke et al. 2018), using the MGII MAGs generated from Parks et al., sought to ask similar questions to the ones I was interested in. Specifically, how many phylogenetically distinct clades of MGII could be determined from the Tara Oceans MAG datasets? And could functional metabolic interpretation provide insights in to how the two major lineages of the MGII – MGIIa and MGIIb – were ecologically distributed? In a testament to the robustness of the methodologies and conclusions, the parallel efforts drew similar/identical conclusions for many of the findings.
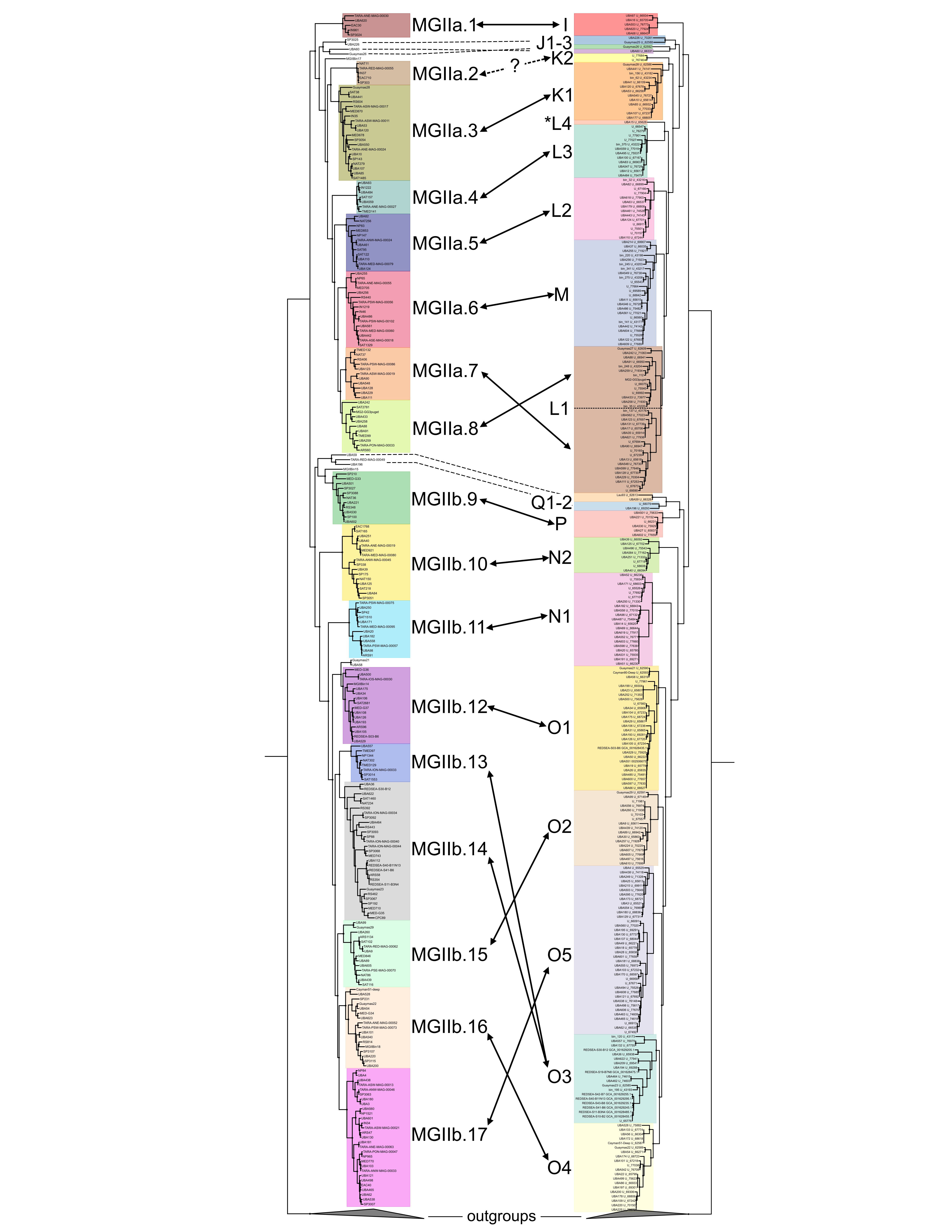
Collectively, the combined work redefines the existing MGIIa (Candidatus Poseidoniaceae) and MGIIb (Candidatus Thalassarchaeaceae) subdivisions – identifying clear phylogenetic subclades, approximately equivalent to genera, that possess metabolic signatures that provide insights into the ecology of the MGII. The MGIIa have the capacity to degrade algal-derived oligosaccharides, possess an archaeal flagellum operon, and are more abundant in coastal regions (or associated with phytoplankton blooms), implicating them as potential opportunistic heterotrophs capable of exploiting ephemeral, biomass rich conditions. In contrast, while the MGIIb maintain the core MGII traits of protein and lipid degradation, they are distributed across the oligotrophic regions of the oceans; dominant in samples lacking their ‘coastal cousins’. The MGII represent an important conduit for marine organic matter degradation and this research will help guide future research efforts to more completely understand this clade of mesophilic archaea.
For more information about the MGII, please find the full manuscript here:
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in