Exploring the CryoPPP Dataset
Published in Research Data

In this blog post, we share our journey and the story behind the paper “A large expert-curated cryo-EM image dataset for machine learning protein particle picking”, that was recently published in Nature Scientific Data. It covers the entire trajectory from conception to publication, it includes the highs and lows, and it describes the challenges we have faced on the way.
 Figure: Overview of Cryo-EM pipeline, from sample preparation to particle recognition.
Figure: Overview of Cryo-EM pipeline, from sample preparation to particle recognition.
Context:
The 3D structures of proteins are important for understanding their interactions with ligands, which enables structure-based drug discovery. The devastating consequences of the COVID pandemic have highlighted the critical importance of expediting the computational-based drug, which is possible after knowing the 3D structures of proteins and complexes.
One of the most powerful techniques for determining the structures of proteins is Cryo-Electron Microscopy (cryo-EM). A key step in constructing 3D protein structures from cryo-EM data is to pick protein particles in cryo-EM images (micrographs), which is a daunting task. The picking task is challenging due to several factors, including high noise levels caused by ice and contamination, low contrast of particle images, particles with heterogenous conformations, and unpredictability in an individual particle’s appearance caused by variation in orientation. Despite these challenges, we recognized and picked protein particles from 34 representative protein datasets. The dataset is 2.6 terabytes and includes 9,893 high-resolution micrographs with labelled protein particle coordinates. It was rigorously validated through 2D particle class validation and 3D density map validation with the gold standard.
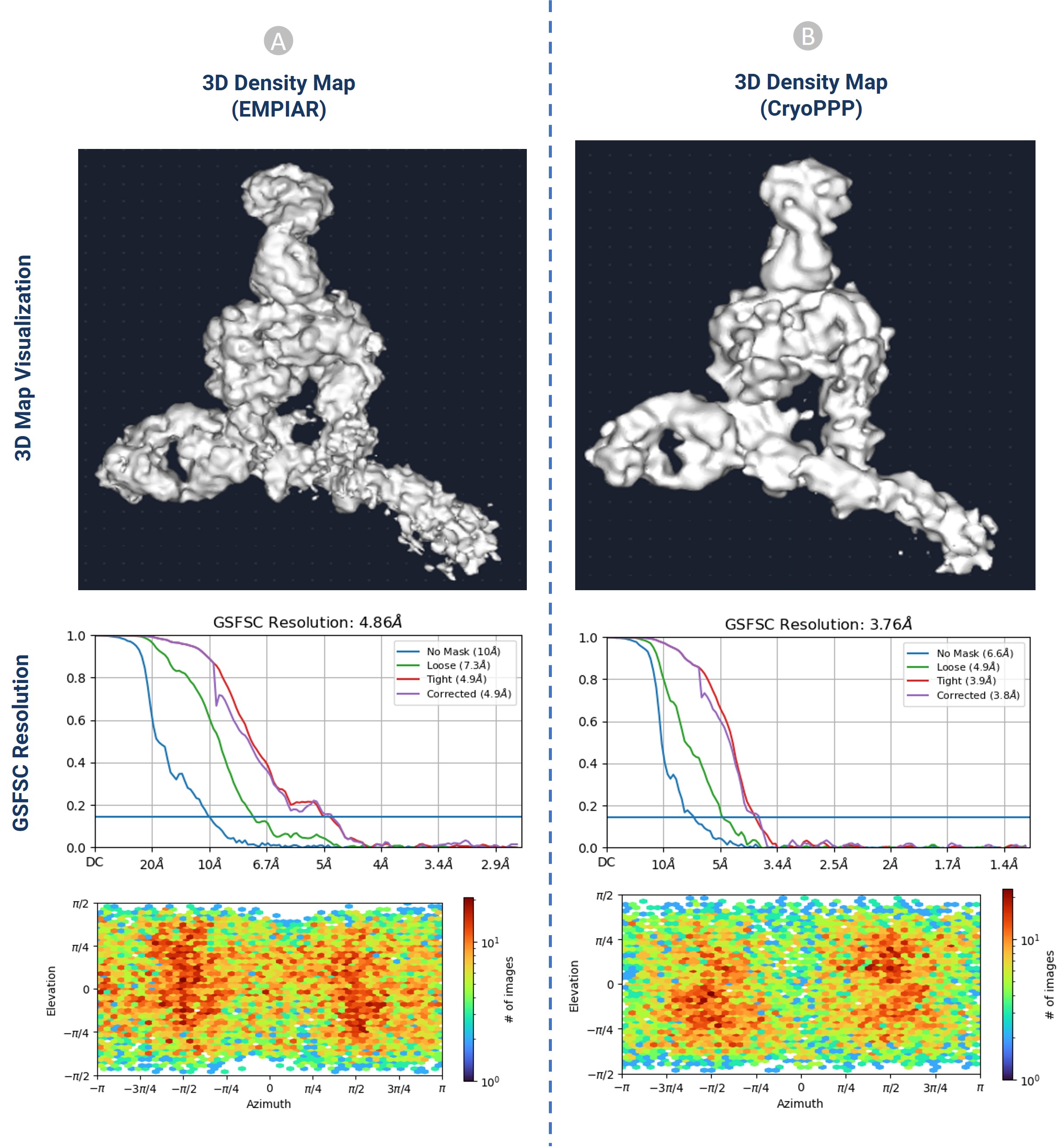
Figure: 3D density map validation of CryoPPP protein particles with EMPIAR protein particles
Exploring the dataset:
The CryoPPP dataset consists of manually labelled 9,893 micrographs of 34 diverse, representative cryo-EM datasets of 34 protein complexes selected from EMPIAR. Each EMPIAR dataset identified by a unique EMPIAR ID has about ~300 cryo-EM images in which the coordinates of protein particles were labeled and cross-validated by two experts aided by software tools.
The full dataset is available at https://github.com/BioinfoMachineLearning/cryoppp. For researchers who have limited disk space, a much smaller light version of CryoPPP, called CryoPPP_Lite, can also be downloaded from the website. CryoPPP_Lite includes the micrograph files in the 8-bit JPG format and the particle ground truth files that only need 121 GB of disk space in total, which is easier to store and transfer.
Each of the data include:
- Raw micrographs: It contains the two-dimensional projections of the protein particles in different orientations stored in different image formats (MRC, TIFF, EER, TIF, etc.)
- Motion correction (gain files): It is used to correct both global motion (stage drift) and local motion (beam-induced anisotropic sample deformation) that occur when specimens (protein particles) are exposed to the electron beam during imaging.
- Particle stack: It comprises of the .mrc files of manually picked protein particles.
- Ground truth label: It contains the star and CSV files for both all true particles (positives) and some typical false positives (e.g., ice contaminations, aggregates, and carbon edges).
What’s Next:
We believe that the CryoPPP dataset would bridge the gap between the computational potential of Deep Learning and the standard benchmarking dataset inadequacy to greatly facilitate the development of AI-based methods for automated cryo-EM protein particle picking. We already started working on pushing the boundaries in this field. Check our recent projects: CryoTransformer and CryoSegNet.
If this sounds as exciting to you as it does to us, we encourage you to download the data set and start exploring it right now!
(Images © Dhakal et al., 2023)
References
- Dhakal, A., McKay, C., Tanner, J. J., & Cheng, J. (2022). Artificial intelligence in the prediction of protein–ligand interactions: recent advances and future directions. Briefings in Bioinformatics, 23(1), bbab476.
- Dhakal, A., Gyawali, R., Wang, L., & Cheng, J. (2023). A large expert-curated cryo-EM image dataset for machine learning protein particle picking. Scientific Data, 10(1), 392.
- Dhakal, A., Gyawali, R., Wang, L., & Cheng, J. (2023). CryoTransformer: A Transformer Model for Picking Protein Particles from Cryo-EM Micrographs. bioRxiv.
- Gyawali, R., Dhakal, A., Wang, L., & Cheng, J. (2023). Accurate cryo-EM protein particle picking by integrating the foundational AI image segmentation model and specialized U-Net. bioRxiv, 2023-10.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in