Extended-spectrum beta-lactamase-producing Enterobacterales: almost a decade persisting in our body
Published in Microbiology
Conception of the study
This study was conceived within the framework of a bigger project designed to investigate the routes of transmission of extended-spectrum beta-lactamase-producing Enterobacterales (ESBL-PE) in both community and hospital settings1, 2, 3, 4. For that, not only isolates retrieved from patients were included, but also environmental isolates recovered from foodstuff (to uncover possible transmission from food to humans) and wastewater (to sense the ESBL-carriage extent in the community and hospitals). However, as a basis to establish realistic thresholds for estimating patient-to-patient transmission, we studied the genetic diversity of the ESBL-PE within the same host5, 6. Our main goals were to estimate strain and plasmid persistence, to quantify the genetic changes in isolates of the same strain over time, and to compare colonizing and infecting isolates within the same host.
How was the study performed and which were the main findings?
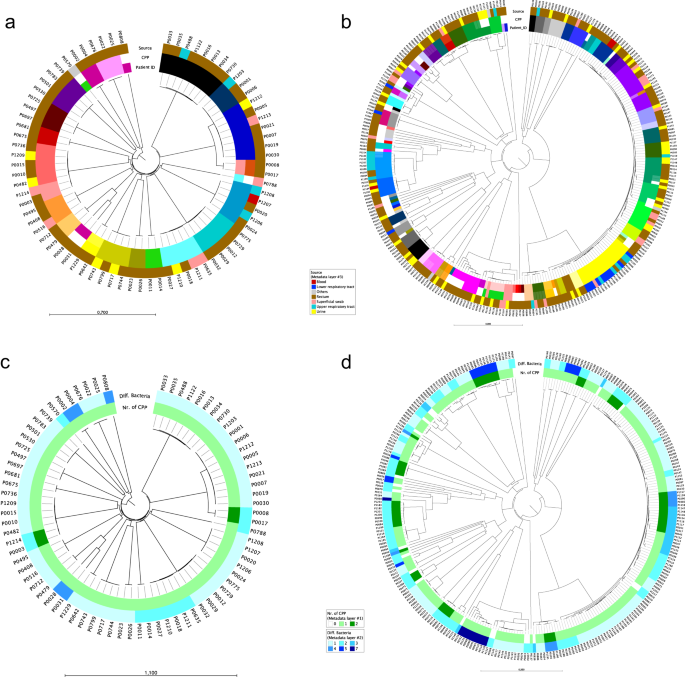
This 10-year longitudinal restrospective study was peformed at the University Hospital Basel, Switzerland. Patients known to be ESBL-PE carriers are screened at each visit to the hospital; this allowed us to build a collection of ESBL-PE over the years. Patients with a at least two consecutive ESBL-PE positive screenings were included in the study.
A total of 76 ESBL-K. pneumoniae species complex and 284 ESBL-E. coli isolates recovered from 19 and 61 patients were analyzed. Persistence of genetically identical strains was demonstrated in the majority of patients. Patients were colonized with the same strain of K. pneumoniae- or E. coli for almost 5 and 9 years, respectively. The number of single nucletide polymorphisms (SNP) differences in subsequent isolates of the same strain with regard to the first isolate of the patient remains almost stable, with a median of 7 SNPs in a median time of 315 and 233 days in K. pneumoniae and E. coli, respectively. Antimicrobial resistance genes, plasmid replicons, and whole ESBL-plasmids were shared between isolates colonizing the same patient, regardless of chromosomal relatedness. In patients with isolates recovered from different body sites, the same strain was frequently found at different sites and related to both infection and colonization.
Main limitations
Since this was a retrospective study, screening intervals were not standardized, resulting in a different number of samples collected at different time intervals for each patient. This hampered a standardized approach to estimating mutation events of colonizing strains over time. Detailed information about antimicrobial use in the study populaton over time was incomplete, thus associations between antimicrobial exposure and genetic diversity could not be investigated. Non-ESBL-PE were not collected, so that the potential loss of plasmids harboring ESBL-genes could not be addressed.
Impact and strenghts
The main strength of this study lies in its longitudinal design of a period of ten years and the high number of samples analyzed for some patients allowing to monitor genetic diversity of strains over a long time period and at different body sites. A combination of different sequencing platforms producing short- and long-reads allowed us to generate closed assemblies of chromosomes and plasmids for many of the isolates included in the study. Hence, we could corroborate the persistence of whole ESBL-plasmids within the same host at different levels (same strain, different strains and even different species). Our finding that isolates belonging to the same strain were observed in the contexts of infection and colonization suggests that host rather than bacterial attributes could be decisive for transition from colonization to infection. In conclusion, the study highlights the importance of long-term persistence of ESBL-PE in individual patients and highlights their role as reservoirs for ongoing transmission and the potential for recurrent infections with colonizing strains.
These results on the bacterial genetic diversity to be expected within individual patients over time, may serve as a valuable basis for further studies designed to analyze strain and host factors related to duration of colonization and transition from colonization to infection in ESBL-E. coli and ESBL-K. pneumoniae complex.
REFERENCES
- Stadler T, et al. Transmission of ESBL-producing Enterobacteriaceae and their mobile genetic elements-identification of sources by whole genome sequencing: study protocol for an observational study in Switzerland. BMJ Open 8, e021823 (2018).
- Gomez-Sanz E, et al. Extended-spectrum beta-lactamase-producing Enterobacterales in diverse foodstuffs: a prospective, longitudinal study in the city of Basel, Switzerland. Front Microbiol 14, 1295037 (2023).
- Gomez-Sanz E, et al. Spatiotemporal dissemination of ESBL-producing Enterobacterales in municipal sewer systems: a prospective, longitudinal study in the city of Basel, Switzerland. Front Microbiol 14, 1174336 (2023).
- Aguilar-Bultet L, et al. Identification of a Cluster of Extended-spectrum Beta-Lactamase-Producing Klebsiella pneumoniae Sequence Type 101 Isolated From Food and Humans. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 73, 332-335 (2021).
- Stoesser N, et al. Extensive within-host diversity in fecally carried extended-spectrum-beta-lactamase-producing Escherichia coli isolates: implications for transmission analyses. Journal of clinical microbiology 53, 2122-2131 (2015).
- Gorrie CL, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 65, 208-215 (2017).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in