Fetal Ultrasound Grand Challenge ISBI 2025
Published in Bioengineering & Biotechnology
Contact: fugc.isbi25@gmail.com
Transvaginal ultrasound is the preferred method for visualizing the cervix in most patients, offering detailed insight into cervical anatomy and structure. Accurate segmentation of ultrasound (US) images of the cervical muscles is essential for analyzing deep muscle structures, assessing their function, and monitoring treatment protocols tailored to individual patients.
The manual annotation of cervical structures in transvaginal ultrasound images is labor-intensive and time-consuming, limiting the availability of large labeled datasets required for robust machine learning models. In response to this challenge, semi supervised learning approaches have shown potential by leveraging both labeled and unlabeled data, enabling the extraction of useful information from unannotated cases. This method could reduce the need for extensive manual annotation while maintaining accuracy, thus accelerating the development of automated cervical image segmentation systems. The envisioned impact of this challenge is twofold: improving clinical decision-making through more accessible and accurate diagnostic tools and advancing machine learning techniques for medical image analysis, particularly in resource-constrained environments.

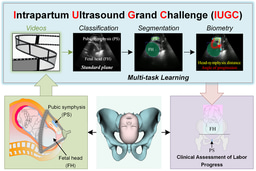
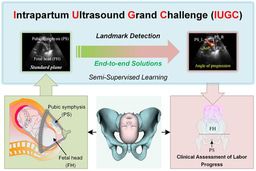
We extend the MICCAI PSFHS 2023 Challenge and the MICCAI IUGC 2024 Challenge from fully supervised settings to a semi-supervised setting that focuses on how to use unlabeled data.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in