Finding Cellular “Zebras” in Type 1 Diabetes Using a Single-Cell Multi-omics Approach
Published in Healthcare & Nursing

Post written by: Maria Fasolino, Gregory W. Schwartz, and Golnaz Vahedi
American medical students have been taught for decades that “when you hear hoofbeats, think of horses, not zebras,” which was coined in the 1940s by Dr. Theodore Woodward. This aphorism is a variation of Occam’s razor, which is a principle that gives preference to simpler hypotheses over the more complex. And thus, this phrase encourages medical trainees to first entertain diagnoses that are simpler or more common, and thus more likely, over those that are more complicated or rare, and thus less probable – which is in reference to the fact that in the United States, horses are more commonplace than zebras. That said, although rare, “medical zebras” do exist, and if sufficient evidence is gathered and a black and white striped pattern is metaphorically emerging, a rare disease or condition may be the appropriate diagnosis.
A similar phrase for type 1 diabetes (T1D) based on its hallmark features and the predominant focus of prior publications would be “when exploring the cellular and molecular immunopathogenesis of T1D, think of beta and immune cells, not other cell types.” However, T1D is an autoimmune disease of only partially defined etiology in which immune cells destroy insulin-producing beta cells, with evidence mounting that other cell types may also contribute. Thus, despite numerous studies on beta and immune cells in T1D, a complete understanding of the etiology of T1D remains unknown. With the advent of single-cell technologies that molecularly characterize enormous numbers of individual cells across multiple modalities, less obvious and/or rare cell types or states are emerging as key players in various diseases and conditions. Motivated by such studies, we sought to determine whether “cellular zebras” exist in T1D – but to do so, we had to employ the most relevant tissue samples for such an analysis.
Historically, most T1D studies have been performed on blood leukocytes, rather than the pancreas, given the inability to safely perform a biopsy of the pancreas in patients and the low beta cell count at the point of T1D diagnosis. Therefore, the etiology of T1D has remained elusive in part due to the inability to molecularly assess cell types associated with disease etiology and sequelae in the site of pathogenesis. This limitation has recently been addressed by a collaboration between the Network for Pancreatic Organ donors with Diabetes (nPOD; https://www.jdrfnpod.org) and the Human Pancreas Analysis Program (HPAP; https://hpap.pmacs.upenn.edu) in which these organizations procure and characterize pancreatic tissues of deceased organ donors that have been recently diagnosed with T1D. Additionally, given that many individuals with T1D harbor pancreatic islet autoantibodies in their bloodstream before clinical diagnosis, nPOD and HPAP also collect samples from donors with islet antibodies but without a medical history of T1D, in hopes of (1) elucidating early events that trigger pathogenesis, and (2) when compared to T1D samples, identifying pathogenic events specifically related to the diabetic state, rather than islet autoantibody positivity.
Therefore, we took advantage of the gold-standard pancreatic datasets generated by the Human Pancreas Analysis Program (HPAP) in three relevant donor groups: individuals with T1D (T1D donors), those with islet autoantibodies but no clinical diagnosis of T1D (AAb+ donors), and those with neither islet autoantibodies nor a history of T1D (control donors). We employed an unbiased, multidimensional approach on an exceptional number of cells and assays: approximately 80,000 cells using single-cell transcriptomics; 7,000,000 cells using cytometry by time of flight (CyTOF); 1,000,000 cells using imaging mass cytometry (IMC); and immunofluorescence staining in native pancreatic tissues.
Armed with these comprehensive, pancreatic molecular datasets, we developed an integrative, quantitative strategy to assess pancreatic islets, systematically identifying cell types and states (please see Figure 1 of the paper). Among several findings describing the pancreatic environment (please see Figures 2 & 3 of the paper), we observed that cells of the exocrine compartment show transcriptional and gene ontological changes in the T1D disease setting (please see Figures 4 to 6 of the paper). Specifically, a subset of ductal cells enriched in T1D donors expressed high levels of MHC class II and interferon pathways. We confirmed the existence of this MHC class II-expressing subset of ductal cells across multiple independent and quantitative experimental assays: scRNA-seq, CyTOF, IMC, and immunofluorescence (see image below, which is from Figure 5i of the paper).
In summary, given that sufficient evidence was gathered, and a black and white striped pattern was metaphorically corroborated across all assays, we posit this MHC class II-expressing subset of ductal cells as “cellular zebras” in T1D. To enable the community to easily explore this cellular population, we created an interactive data explorer that provides direct access to the single-cell transcriptomics data presented in the paper (https://cellxgene.cziscience.com/collections/51544e44-293b-4c2b-8c26-560678423380/).
Through the usage of the optimal biological material available and high throughput multimodal analyses, we were able to delineate cell types and processes that may contribute to T1D immunopathogenesis and provide an integrative approach for the exploration of human pancreatic function. This comprehensive profiling of cellular states in T1D allowed for the identification of “cellular zebras” in this disease setting, serving as a framework for the discovery of “cellular zebras” using single-cell multi-omics data in additional disease states and conditions.
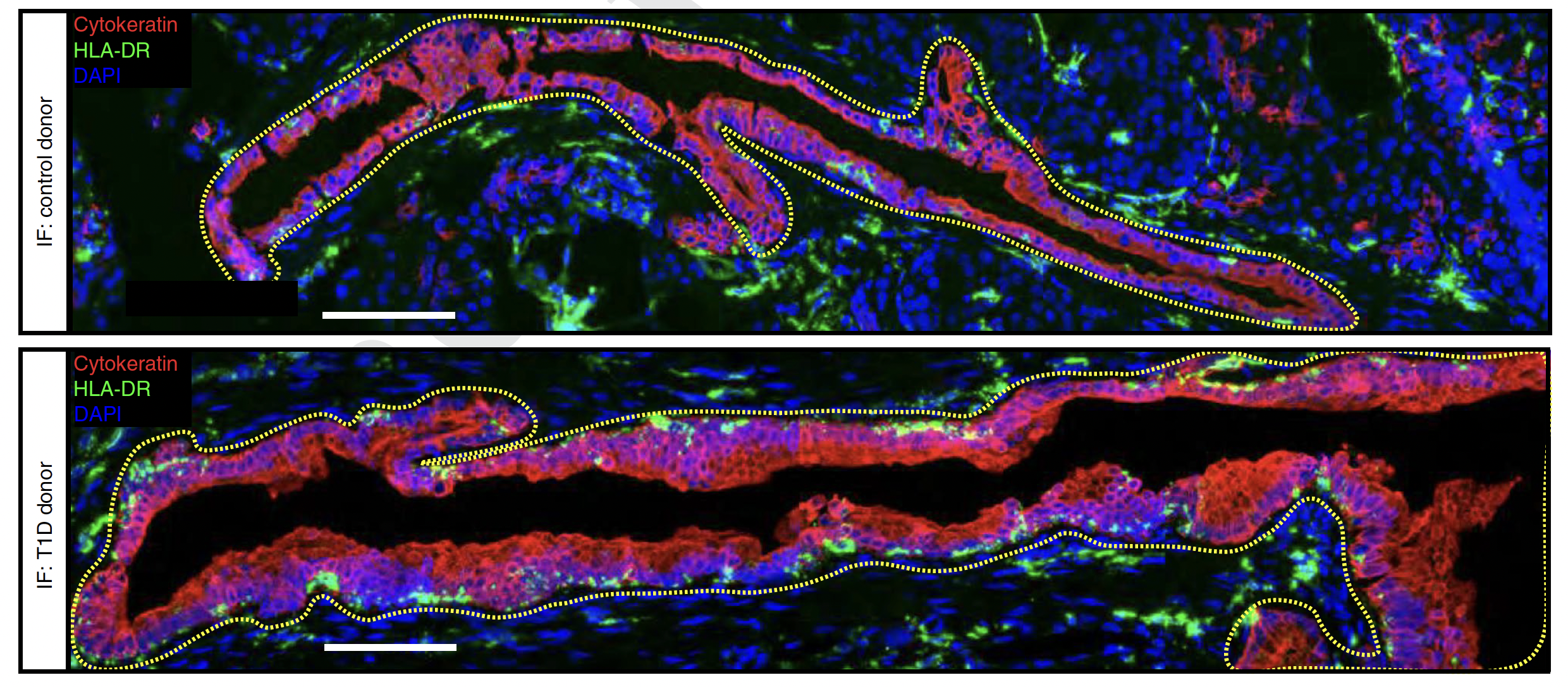 From Figure 5i of the paper: Representative confocal microscopy images from the pancreas of a control donor (top) and T1D donor (bottom) displaying HLA-DR+ Cytokeratin+ cells labelled by immunofluorescence (IF; control, n = 3; T1D, n = 2; Cytokeratin is a ductal cell marker; HLA-DR is an MHC Class II cell surface receptor).
From Figure 5i of the paper: Representative confocal microscopy images from the pancreas of a control donor (top) and T1D donor (bottom) displaying HLA-DR+ Cytokeratin+ cells labelled by immunofluorescence (IF; control, n = 3; T1D, n = 2; Cytokeratin is a ductal cell marker; HLA-DR is an MHC Class II cell surface receptor).
Access the paper here: https://www.nature.com/articles/s42255-022-00531-x
Follow the Topic
-
Nature Metabolism

This journal publishes work from across all fields of metabolism research that significantly advances our understanding of metabolic and homeostatic processes in a cellular or broader physiological context, from fundamental cell biology to basic biomedical and translational research.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in