Finding the microbial heroes: Identifying the smallest groups of microbes, capable of executing a desired function, from a large microbial community
Published in Protocols & Methods, Anatomy & Physiology, and Mathematics
The mammalian gut is home to at least 1000 different microbial species1 that play a crucial role in the overall health of the host. These microbes co-exist as a community in the gut and flourish by exchanging metabolites. Their role in aiding digestion is non-trivial. The breakdown of dietary fibres into products absorbable by the human body is achieved only with their assistance. The gut microbiome produces short-chain fatty acids (SCFAs) that are known to promote health and modulate inflammatory responses. The gut microbiome impacts even distant organs such as the liver, lungs and heart. Recent studies have shown that the gut microbiome also influences cognition and emotional behaviours due to the bidirectional relationship along the brain-gut-microbiome axis. Disruption of the gut microbiome has been found to be associated with a number of disease states, such as obesity, type II diabetes, inflammatory bowel disease, some types of cancer, etc.2
Disruption of the gut microbiome may be caused by several factors such as antibiotic usage, alcoholism, chronic stress, changes to circadian rhythm, altered diet, etc. Any such perturbation to the gut microbiome can impact the overall health of a person. Although consuming probiotics and prebiotics can help replenish the gut microbiome, in some cases, it may be more effective to reintroduce the lost flora to the gut. For instance, some cases of severe IBD (Inflammatory Bowel Disease) and Clostridium difficile infections are treated using the faecal transplant strategy, which is transferring a portion of the faecal matter of a healthy donor to the patient’s gut. However, this strategy comes with a high risk of transmitting any pathogens that may be present in the donor’s gut. A safer treatment strategy is to artificially build a synthetic microbial community of known composition, with the required functionality, to be transplanted into the patient’s gut. Then, the interesting question arises - how do we decide on the composition of such a synthetic community?
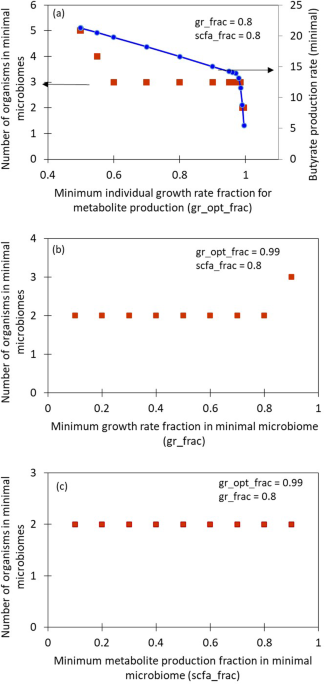
Consider a functionality, such as the overall butyrate production of the large microbial community: all the members of the community do not contribute equally to it. It is possible to find a smaller subset of species that can achieve this functionality while maintaining the required community growth rate. We present an algorithm to identify such subsets of a given large microbial community that is present in the healthy donor. We call this subset, a minimal microbiome. A minimal microbiome will consist of the keystone species for performing the functionalities of interest. For instance, a butyrate-producing minimal microbiome may contain at least one Firmicute species, which are known to be butyrate producers. Production of multiple metabolites together and alone are the functionalities considered in the model. Because multiple organisms are capable of similar functionalities due to functional redundancy, it is possible to find multiple minimal microbiomes for the same desired functionality. The number and type of species in a minimal microbiome are dependent on the community features, such as minimum overall growth rate, that can be set by the user using the tunable parameters in the algorithm. Analysis of different minimal microbiomes obtained from some model gut microbial communities is presented in the paper. A quantitative, constraint-based, flux balance analysis (FBA) approach is used in this model and is implemented using COBRA toolbox3. In order to find the minimal microbiome, a top-down approach of sequential deletion of species followed by mixed integer linear programming (MILP), to minimize the L1 norm of the membership vector is employed. The sequential deletion can be in a user-specified order or a random order that the model can generate. This step can help reduce the computational difficulty of solving a large MILP, in addition to helping to find a variety of minimal microbiomes. The algorithm can identify different minimal microbiomes if present and report their growth rate and metabolite production rates. The workflow of the algorithm is presented in Figure 1.
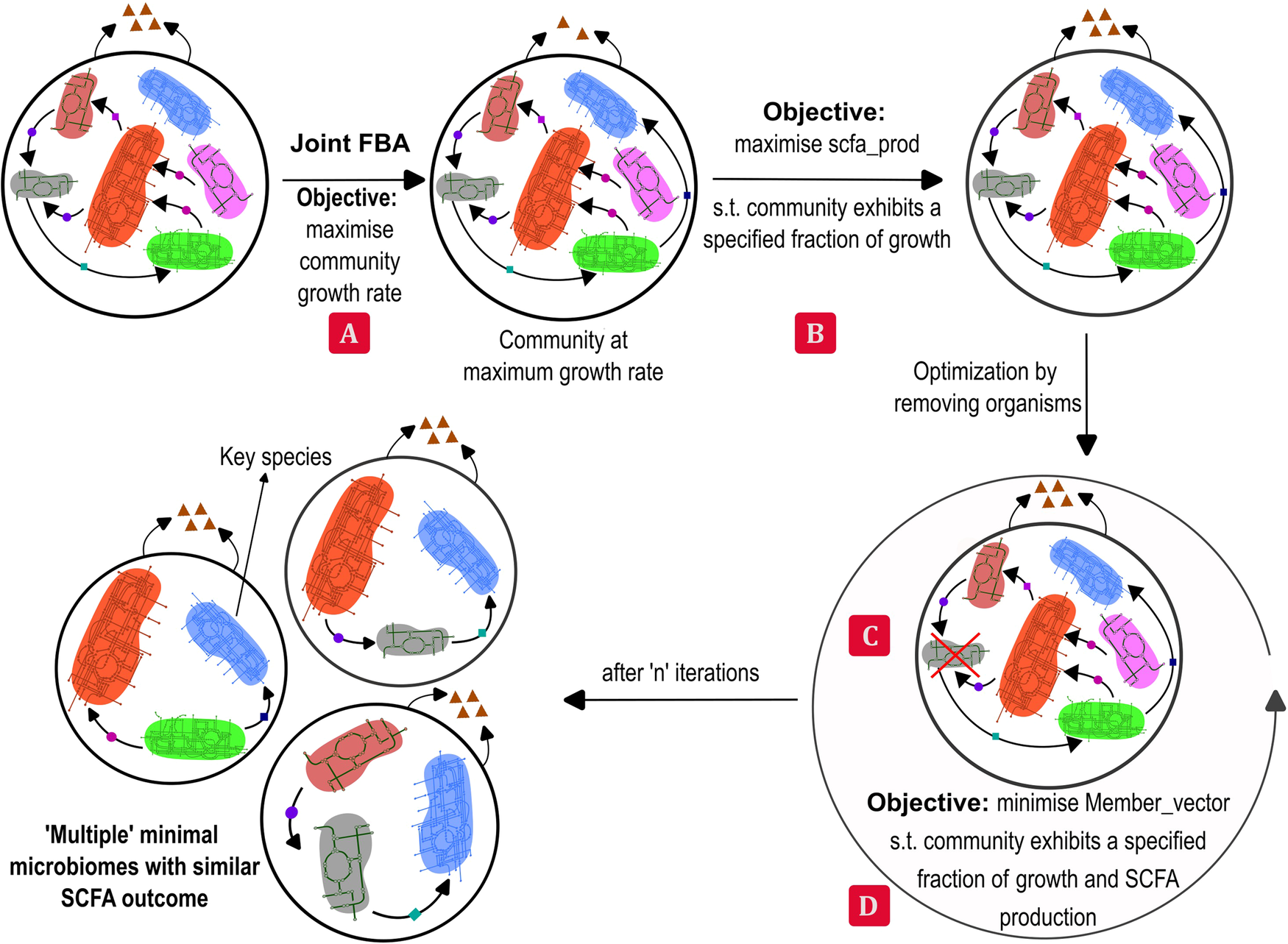
Figure 1: Workflow of the minMicrobiome algorithm
This algorithm will be useful to design personalized treatment strategies for diseases caused by perturbations to the gut microbiome. Although presented as a method to work with the gut microbiomes, this approach is generic and flexible and can be extended to use in the case of other co-operative microbial communities such as those used in bioreactors. The algorithm is available from https://github.com/RamanLab/minMicrobiome.
References
- Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, et al. A framework for human microbiome research. Nat 2012 4867402. 2012;486:215–21.
- Hollister EB, Gao C, Versalovic J. Compositional and Functional Features of the Gastrointestinal Microbiome and Their Effects on Human Health. Gastroenterology. 2014;146:1449–58.
- Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc. 2019;14:639–702.
Follow the Topic
-
npj Systems Biology and Applications

An online Open Access journal dedicated to publishing the premier research that takes a systems-oriented approach and encourages studies that integrate, or aid the integration of, data, analyses and insight from molecules to organisms and broader systems.
Related Collections
With Collections, you can get published faster and increase your visibility.
Next-Generation Mammalian Cell Bioprocessing: Systems Biology, Synthetic Biology, and Beyond
Publishing Model: Open Access
Deadline: Mar 15, 2026
Systems mechanobiology
Publishing Model: Open Access
Deadline: Mar 28, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in