From noise to signal, how we flipped a problem into a paper
Published in Ecology & Evolution

We recently published a paper where we use a new approach to study the impact of variation in both chloroplast and mitochondrial genomes on plant phenotypes. Assessing the impact of variation in these organellar genomes (collectively referred to as the plasmotype and the nuclear genome as the nucleotype) on phenotypes has always been tricky, primarily because it is not always easy to differentiate the impact of plasmotypic variation from variation in the nucleotype. Over the years various strategies have been used such as reciprocal cross designs, repeated backcrossing, and even protoplast fusion to physically exchange plasmotypic and nucleotypic genomes. Each of these approaches has limitations and pitfalls and a precise assessment of the phenotypic impact of plasmotypic variation on plants has largely eluded the research community. We developed a simple approach which allows exchange of nucleotype and plasmotype in as little as a single generation. This not only allows a detailed investigation of how variation in the plasmotype effects plant performance and fitness but also allows modified organelles to be transferred to new nucleotypic backgrounds. Plant organelles are notoriously difficult to transform and this approach offers a valuable way to amplify successful transformation events and transfer them into elite germplasm. However, when we started our PhDs none of this was on our minds.
Back in 2010 a paper was published by Ravi Maruthachalam and Simon Chan where they described a plant expressing a modified cenh3 protein which caused uniparental genome elimination resulting in haploid offspring. At the same time, I had just run my first experiments on a high throughput phenotyping system capable of measuring 1440 plants in under an hour for photosynthetic efficiency. Once the system was properly working one of the first things we did was a genome wide association study (GWAS), which was new and flashy at the time, for photosynthesis. Once the first dataset came in the most striking result was that one genotype (Ely) was very different from the others. This genotype was collected from a railway track in England where they were spraying a lot of triazine herbicides and in response to this herbicide resistance had evolved. This was interesting enough on its own that it led to a whole other research project. BUT it totally messed up my GWAS. The Ely genotype had a very distinct photosynthetic phenotype which was genetic in origin, the only problem was that the causal mutation was in the chloroplast genome yet all my GWAS markers were on the nuclear genome. When Ely was included in the GWAS it resulted in a bunch of false positives, something I termed ‘cytoplasmic noise’. This worried me as I was interested in photosynthesis and wondered how much of the variation was nuclear in origin and how much was coming from variation in the mitochondria or chloroplast genomes, the plasmotype.
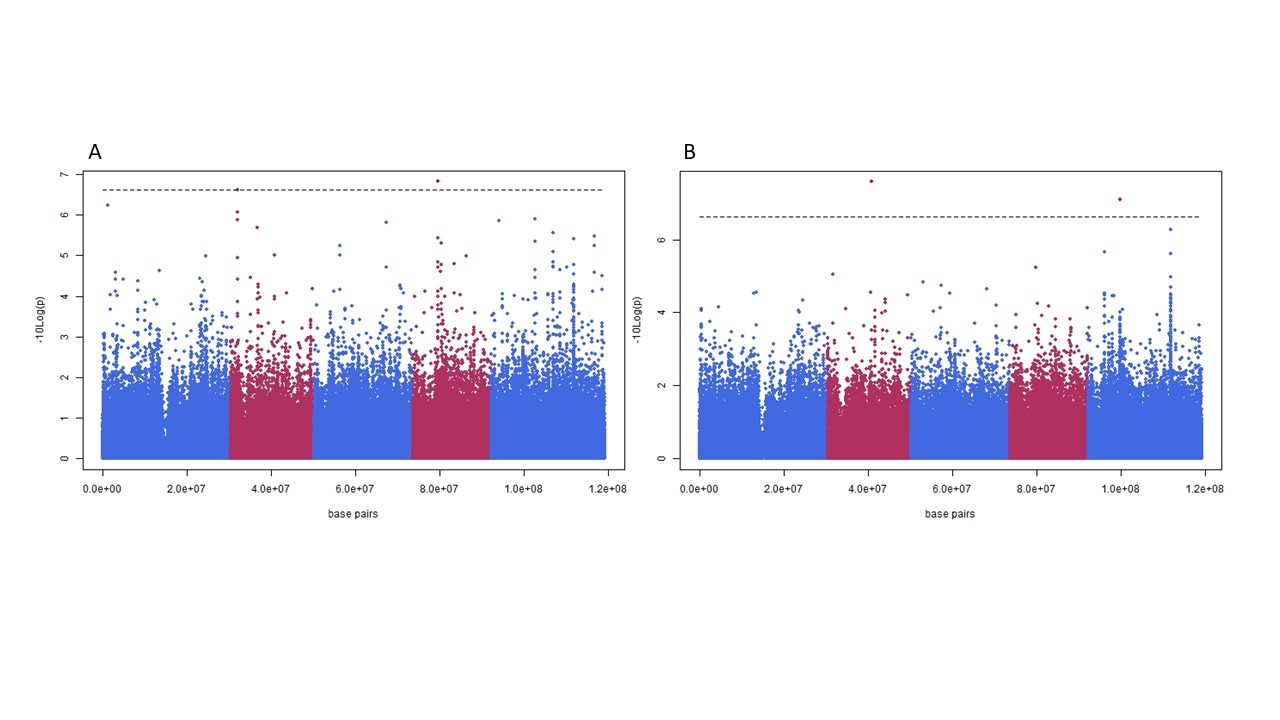
The impact of large effect variants in the organelles on a phenotype can results in false positive associations, “cytoplasmic noise”. Panel A shows a Manhattan plot for photosynthetic efficiency in a panel of 360 Arabidopsis thaliana accessions. Panel B is the same plot with a single genotype removed, this genotype contains a chloroplast encoded mutation which has a large impact on photosynthetic efficiency, large enough that is drives spurious associations to non-causal nuclear loci.
I presented my concerns in a departmental seminar in summer 2010 where a fellow PhD student Erik Wijnker was in the audience. He collaborated with Ravi and Chan for his studies in plant meiosis and realised I might be able to use the cenh3 haploid inducer to solve the problem of “cytoplasmic noise”. We developed the idea over a few beers and decided to go ahead and try it out. It was not so simple since we were both just PhD students, but we were able to sell the idea and enlist the help of many enthusiastic researchers, as can be seen from the author list. By the end of my PhD I had generated a lot of data and written a thesis chapter but the project was far from complete. Both myself and Erik were pursuing postdocs elsewhere. At this point Tom Theeuwen got in touch and said he would like to follow up on this research for his PhD. Tom brought fresh energy to the project and managed to tame what was a large and unwieldly dataset into something coherent. Even so it still took us some time to properly get our heads around the data. For me one of the most fun insights is that variation in the plasmotype may act as an evolutionary capacitor. Variation is often only visible in specific nuclear and environmental contexts, as such there is a potential reservoir of adaptive variation waiting to be tapped. So, what at first might be a problem or noise in your data can end up driving some very interesting research, often you just need to talk to somebody who has a different perspective.
Follow the Topic
-
Nature Plants

An online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in