From the BugBitten Archives: Clocking out -knocking out circadian clock gene disrupts key functions in Aedes mosquitoes
Published in Microbiology and Zoology & Veterinary Science

This blog was originally posted on the old BugBitten WordPress site . We are resharing it now on the Research Communities site.
Aedes aegypti is a species of mosquito that is a disease vector for several arboviral diseases such as dengue, Zika and chikungunya that affect millions of people globally. Many of these diseases do not have vaccines readily available, and so a lot of emphasis is put on controlling the spread of the vector. Molecular approaches to vector control have been emerging over the past ten years, with genetically modified mosquitoes being released into the wild to control breeding in mosquito populations.
Vinaya Shetty and colleagues have been looking at another way to disrupt mosquito populations, by investigating whether removing key genes related to the circadian clock of the mosquitoes could impact their clock-dependent biological processes.

Mosquitoes display time-related behaviours such as feeding at certain times of the day. For A. aegypti feeding is a day time activity – mainly in the early morning and late afternoon – whilst the anopheline mosquito is a nocturnal feeder.
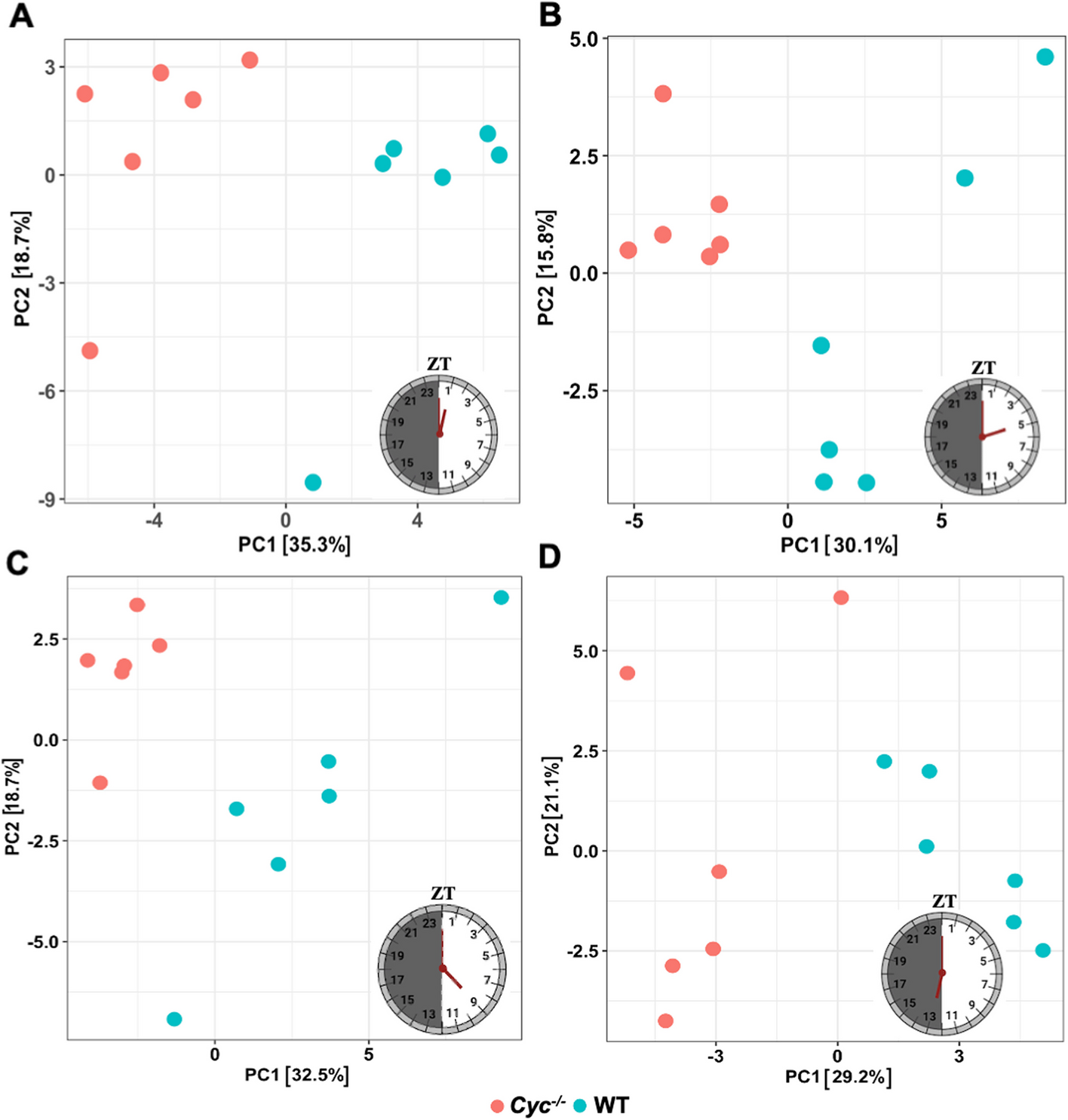
In their 2022 study, Vinaya Shetty and colleagues knocked out the core clock cycle gene in A. aegypti using CRISPR/Cas9 and found the mRNA expression of 7 circadian related genes were changed. This knock out affected the mosquito’s longevity, feeding pattern and reproductive fitness. Their latest study explored this further by using transcriptome profiling and differential gene expression to determine the network of genes and biological pathways that were affected by disrupting the clock gene.
Differential gene expression enabled Shetty and colleagues to identify thousands of genes that were expressed differently in day-night cycles, and determine that removal of the cycle gene caused disruption in metabolic processes, signaling pathways and immune responses. This showed the vital role that the circadian clock plays in A. aegypti’s life, and targeting it could see massive changes to the mosquito’s behaviour and consequently neutralise its impact as a disease vector.
Cover image credit: Gerd Altmann from Pixabay
Follow the Topic
-
BMC Genomics

This is an open access, peer-reviewed journal that considers articles on all aspects of genetics, genomics and proteomics.
-
BugBitten

A blog for the parasitology and vector biology community.
Related Collections
With Collections, you can get published faster and increase your visibility.
Genomics of microbiomes
The study of microbiomes has emerged as a dynamic field at the intersection of genomics, ecology, and health sciences. Microbiomes encompass the diverse communities of microorganisms, including bacteria, viruses, and unicellular eukaryotes, residing in various environments, such as the human body, food, soil, and aquatic systems. Understanding the genomic makeup of these microbiomes is crucial for unraveling their complex interactions with hosts and the environment. As advances in sequencing technologies, including single molecule sequencing, metagenomics and single cell omics, continue to evolve, researchers are better equipped to explore the rich genetic diversity (including pangenomes and epigenomes) and functional capacities of microbiomes across different ecosystems.
Investigating the genomics of microbiomes is pivotal for addressing critical questions in ecology, health, disease, and environmental sustainability. For instance, recent breakthroughs in the field have illustrated how microbiomes influence human health, from their roles in metabolism and immune function to their impact on mental health. Furthermore, understanding the genomics of environmental microbiomes can provide insights into biogeochemical processes and ecological resilience. As we deepen our knowledge of these microbial communities and develop computational biology methods to model their functionality, we stand to enhance our ability to harness their potential for applications in medicine, agriculture, and environmental management.
Future research in this area holds the promise of transformative advances in our understanding of microbiomes. The integration of multi-omics approaches, combining genomics with transcriptomics, proteomics, and metabolomics, may lead to a holistic view of microbial community dynamics and their functional implications. Additionally, developments in artificial intelligence and machine learning could further accelerate discoveries, enabling the identification of novel microbial functions and their roles in health and disease. As we continue to explore these intricate relationships, we can anticipate innovative strategies for harnessing microbiomes for therapeutic and environmental applications.
Topics of interest include, but are not limited to:
•Genomics and epigenomics of host-microbe interactions
•X-omics studies in environmental and host-microbiomes
•Advances in genomics of unculturable microorganisms
•Genome-guided development of synthetic microbiomes and consortia
•Microbiomes and environmental resilience: a genomic perspective
•The human microbiome: genetic diversity and functional potential
•Microbial adaptation and evolution in changing environments
•The role of microbiomes in antibiotic resistance and pathogenesis
•Computational and AI-driven methods for microbiome genomics
•Microbiome applications in sustainable agriculture and environmental management
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 07, 2026
Cattle genomics
BMC Genomics invites researchers to contribute to our Collection on Cattle genomics focusing on understanding the genetic makeup of bovine species, which is essential for improving livestock breeding and health. Advances in genomic technologies, such as next-generation sequencing and RNA sequencing, have enabled researchers to reveal insights into traits such as growth, meat quality, milk production, disease resistance, reproductive fitness, and overall adaptability in bovine genomes. This Collection aims to highlight the latest research developments in cattle genomics, encompassing both genomic and transcriptomic studies that contribute to the understanding of bovine biology.
Recent breakthroughs in genomic selection and precision breeding techniques have already shown promise in increasing efficiency in cattle production. The use of CRISPR-Cas genome editing, for example, has allowed for precise modifications to the cattle genome, introducing beneficial genetic variations without the linkage drag associated with traditional breeding methods. Additionally, the integration of omics technologies is paving the way for a holistic understanding of cattle biology, allowing for more effective management and breeding strategies. Studying the rumen microbiome using genomics, transcriptomics, proteomics, and metabolomics has revealed how microbial communities contribute to feed efficiency and nutrient absorption. This comprehensive approach enables targeted nutritional strategies that improve cattle health and productivity while reducing environmental impact. Such integrative studies facilitate the selection of cattle with optimal microbiome compositions, leading to more sustainable and efficient cattle production systems.
As research in cattle genomics progresses, we can anticipate the development of more sophisticated genomic tools that will enable precise manipulation of genetic traits in bovine populations. This may lead to enhanced resilience against diseases, improved reproductive performance, and better adaptation to changing environmental conditions. Ultimately, continued innovation in this field holds the potential to reform cattle production systems, ensuring sustainable livestock farming for future generations.
- Genomic selection in cattle breeding
- Transcriptomic analysis of bovine traits
- Pathogenicity and disease resistance genomics
- Advances in RNA-Seq applications for cattle
- Omics approaches to cattle health and productivity
- Genetic mapping of economically important traits
- Gene editing
- Metagenomics of the bovine gut microbiome
- Epigenetic regulation of growth and reproduction
- Comparative genomics of cattle and other livestock species
This Collection supports and amplifies research related to SDG 2, Zero Hunger.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: May 26, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in