From the Skies to the Scanner: Precision and Challenges in CT Scanning a Second World War Messerschmitt Me 163
Published in Materials, Research Data, and Computational Sciences
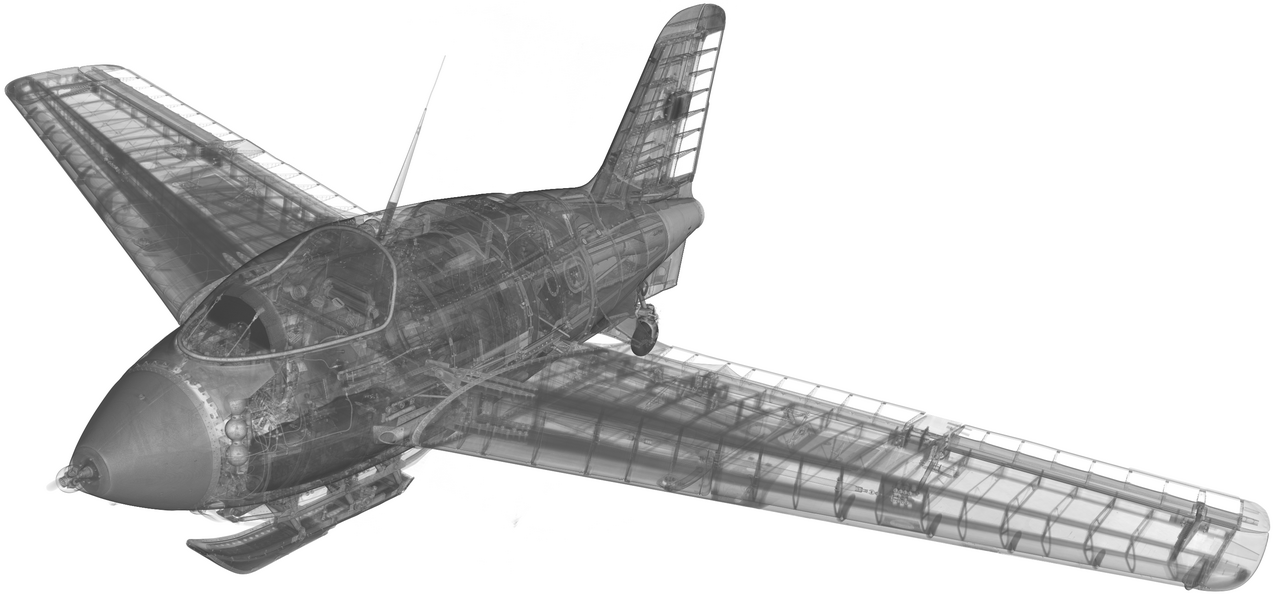
Alright, buckle up and set your seats into an upright position. Today, we’re soaring (this, my seach engine tells me, is another aircraft related pun) into the thrilling world of X-raying and CT-scanning a historic Second World War aircraft. This blog post invites you on an insider’s tour of our publication "Selected annotated instance segmentation sub-volumes from a large scale CT data-set of a historic aircraft". Our team took on the challenging task of manually segmenting and releasing subsets of a CT volume dataset of the Messerschmitt Me 163, a historic German rocket powered interceptor aircraft from the second world war. Join us as we discuss the hurdles we faced and share insights into the dataset that we’ve made available for download . We aim to provide insight into our decision process, explain the reasons for releasing the data, and share some of our concerns about making this information public.
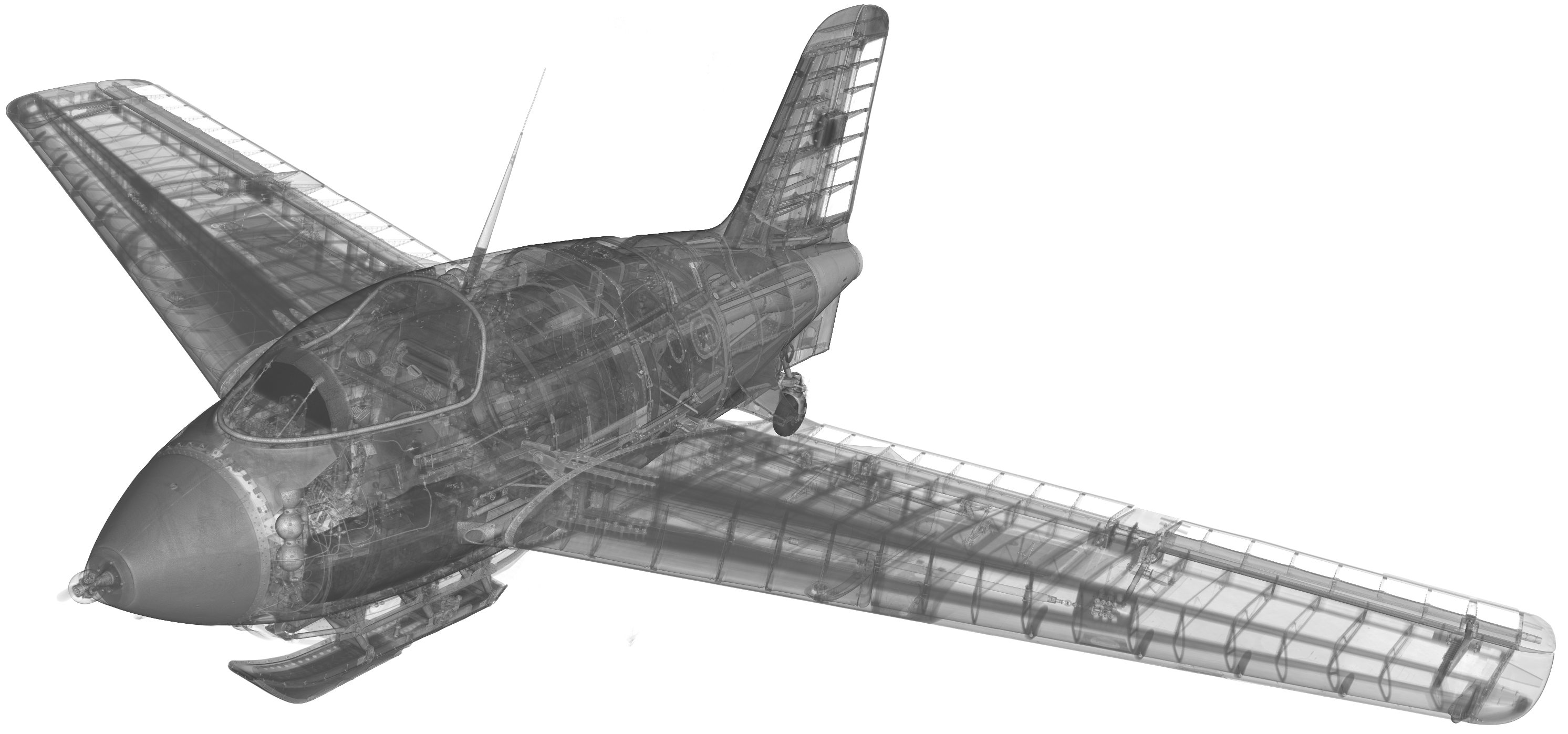
Mission Possible? Aiming for Precision in XXL-CT Aircraft Data
Why do we go to great lengths to X-ray and virtually dissect a historic aircraft? Sure, it’s inherently fascinating by itself. But the primary objective is about gaining scientific insights from a historical viewpoint. The Me 163, displayed at the Deutsches Museum München and accessible to the public, still holds many mysteries. After being captured in 1945 and later modified by the Royal Air Force for test flights, for a long time it served merely as a technological curiosity and only later found its way into the museum. Throughout the years of restoration and maintenance work, most of the maintenance hatches and access points were plastered over and sealed, making them inaccessible without causing damage to the aircraft. Only decades later the arrival of Computed Tomography systems capable of scanning huge objects made it possible to create detailed CT images of objects this size. Originally developed for capturing automotive data, the facility was ideally suited to scan the entire aircraft. Only the wings were temporarily removed and scanned in a second step.

Metal Mayhem: Why Counting Rivets Isn't as Easy as It Sounds! And Why We're Betting on Automatic Segmentation!
Creating the CT volume data (a 3D-grid of voxel values not knowing of each other) was just the beginning or rather, a steppingstone. As detailed as the data are, interpreting them without expertise is no walk in the park. Questions like "Where does this metal sheet end?", "Is this a data acquisition or reconstruction artifact or is something inside the cavity?", or "How many rivets and meters of cable does the aircraft have?" are supposed to be answerable through the CT data but proved to be complex in detail. So, we realized that we need an automatic instance segmentation in the long run.
The Annotator's Marathon: To annotate or not to annotate? Repetition, Injury, Perseverance, and the Questionable Data!
Creating trustworthy validation, test, and training data is one of the first, longest, and crucial steps in segmentation algorithm development. We chose a semi-manual method, annotating seven 512³ large volume blocks from the overall dataset with several annotators. The exact approach and the issues we encountered are described in the publication itself. What we didn’t mention is the mental load weighing on the annotators. Annotating a single plausible volume layer is relatively easy, but ensuring high data quality results requires making many tough decisions consistently across hundreds of layers throughout weeks and months. We spent many hours and discussions ironing out our mistakes and misconceptions and afterwards aligning the annotations with our decisions. Often, we referred to documents about the various manufacturing techniques likely used, as well as macrograph images of rivet connections, to arrive at the segmentation decisions found in the dataset. These decisions were made with the best knowledge and conscience but are certainly not correct everywhere. Besides the mental strain of the annotators, the physical toll also should not be underestimated. During a week with fewer tasks, it might seem tempting to spend extra hours annotating. However, doing so can easily lead to strain on your wrists, eyes, and back. But hey, you can listen to an audiobook or something on the side... the little things in life, right?
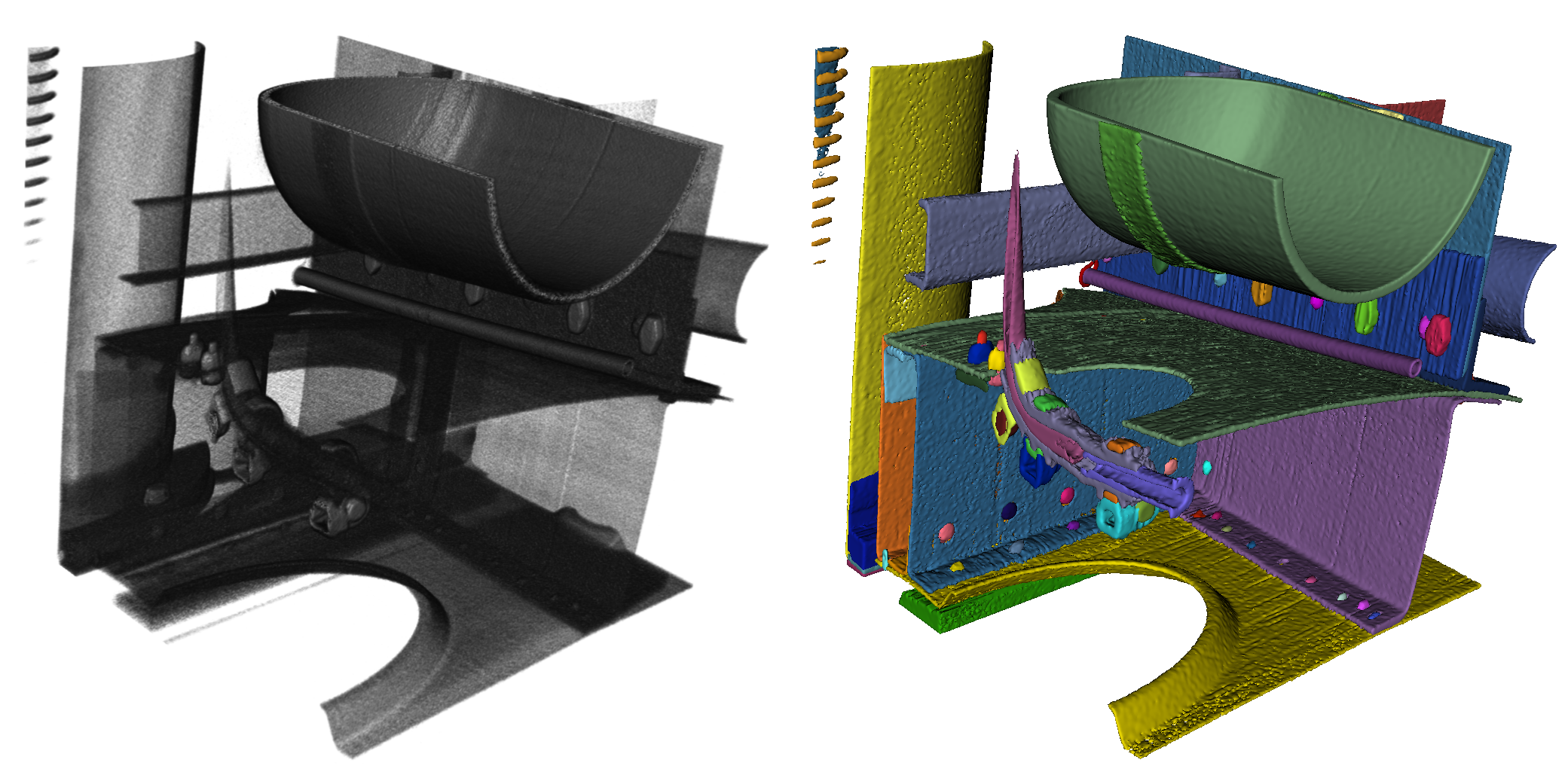
The Fear of a trivial solution and being outsmarted: What If Our Complex Problem Is Just Child's Play?
During and after creating the annotation data, as part of a long-term research project, we further developed methods for as automatically as possible instance segmentation of these and similar datasets (hint: there are quite more automobiles than historic jet planes to be analyzed in the future). Our initial results were promising but not yet reliable enough to segment the aircraft with them. After some experimenting, we decided to launch the “Instance Segmentation XXL-CT Challenge of a Historic Airplane“ with our data, where we published our training data and invited teams worldwide to tackle the segmentation problem with their methods. We were very nervous because we were working on an ongoing research project we had invested a lot of time and resources into. What if someone just wants to snatch the data and disappears? Or, worse, simply solve the problem and make us look incapable? What if it turns out we were just clumsy with our methods? After some internal discussions, we decided that "solving the problem is more important than our ego" and released the data for the challenge.
As the results of the "XXL-CT Segmentation Challenge” gradually became apparent, we felt both relieved and worried. Many registered participants preferred not to submit a segmentation proposal, often because, according to their own statements, they found the problem too difficult or the work effort too high. And the two teams that submitted a segmentation proposal were on a similar level as our own attempts. Phew, relief that we might not have completely botched it. But wait, the worry remains because the problem itself is still unsolved.
This brings us back our publication "Selected annotated instance segmentation sub-volumes from a large scale CT data-set of a historic aircraft". We continue working on the instance segmentation of large-volume CT datasets, but at the same time, publish our data in the hope and fear that someone might tackle the problem with methods unknown to us and publish the progress. We seek to advance together and, in the process, get cited like crazy.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Invertebrate omics
Publishing Model: Open Access
Deadline: May 08, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in